3EHF
 
 | |
5FTU
 
 | | Tetrameric complex of Latrophilin 3, Unc5D and FLRT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADHESION G PROTEIN-COUPLED RECEPTOR L3, CALCIUM ION, ... | | Authors: | Jackson, V.A, Mehmood, S, Chavent, M, Roversi, P, Carrasquero, M, del Toro, D, Seyit-Bremer, G, Ranaivoson, F.M, Comoletti, D, Sansom, M.S.P, Robinson, C.V, Klein, R, Seiradake, E. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.01 Å) | | Cite: | Super-Complexes of Adhesion Gpcrs and Neural Guidance Receptors
Nat.Commun., 7, 2016
|
|
5LTR
 
 | |
8BO3
 
 | | COAGULATION FACTOR XI PROTEASE DOMAIN IN COMPLEX WITH ACTIVE SITE INHIBITOR Asundexian | | Descriptor: | 4-[[(2~{S})-2-[4-[5-chloranyl-2-[4-(trifluoromethyl)-1,2,3-triazol-1-yl]phenyl]-5-methoxy-2-oxidanylidene-pyridin-1-yl]butanoyl]amino]-2-fluoranyl-benzamide, CITRIC ACID, Coagulation factor XIa light chain | | Authors: | Schaefer, M, Roehrig, S, Ackerstaff, J, Nunez, E.J, Gericke, K.M, Meier, K, Tersteegen, A, Stampfuss, J, Ellerbrock, P, Meibom, D, Lang, D, Heitmeier, S, Hillisch, A. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Design and Preclinical Characterization Program toward Asundexian (BAY 2433334), an Oral Factor XIa Inhibitor for the Prevention and Treatment of Thromboembolic Disorders.
J.Med.Chem., 66, 2023
|
|
8BO5
 
 | | COAGULATION FACTOR XI PROTEASE DOMAIN IN COMPLEX WITH ACTIVE SITE INHIBITOR 3 | | Descriptor: | 2-[4-(3-chlorophenyl)-2,5-bis(oxidanylidene)pyrrolo[3,4-b]pyridin-1-yl]-~{N}-[4-(1~{H}-1,2,3,4-tetrazol-5-yl)phenyl]ethanamide, CITRIC ACID, Coagulation factor XIa light chain, ... | | Authors: | Schaefer, M, Roehrig, S, Ackerstaff, J, Nunez, E.J, Gericke, K.M, Meier, K, Tersteegen, A, Stampfuss, J, Ellerbrock, P, Meibom, D, Lang, D, Heitmeier, S, Hillisch, A. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Preclinical Characterization Program toward Asundexian (BAY 2433334), an Oral Factor XIa Inhibitor for the Prevention and Treatment of Thromboembolic Disorders.
J.Med.Chem., 66, 2023
|
|
5I9U
 
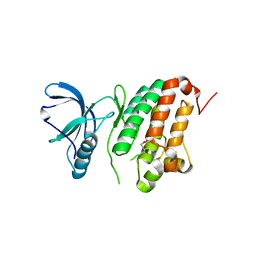 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
8BO6
 
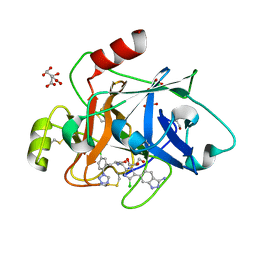 | | COAGULATION FACTOR XI PROTEASE DOMAIN IN COMPLEX WITH ACTIVE SITE INHIBITOR 2 | | Descriptor: | (~{E})-~{N}-[[5-(3-azanyl-1~{H}-indazol-6-yl)-4-chloranyl-1~{H}-imidazol-2-yl]methyl]-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enamide, CITRIC ACID, Coagulation factor XIa light chain, ... | | Authors: | Schaefer, M, Roehrig, S, Ackerstaff, J, Nunez, E.J, Gericke, K.M, Meier, K, Tersteegen, A, Stampfuss, J, Ellerbrock, P, Meibom, D, Lang, D, Heitmeier, S, Hillisch, A. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design and Preclinical Characterization Program toward Asundexian (BAY 2433334), an Oral Factor XIa Inhibitor for the Prevention and Treatment of Thromboembolic Disorders.
J.Med.Chem., 66, 2023
|
|
8BO7
 
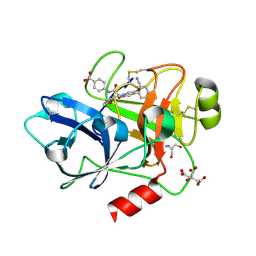 | | COAGULATION FACTOR XI PROTEASE DOMAIN IN COMPLEX WITH ACTIVE SITE INHIBITOR 34 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-[[(2~{S})-2-[4-(5-chloranyl-2-cyano-phenyl)-3-methoxy-6-oxidanylidene-2,5-dihydropyridin-1-yl]-3-[(2~{S})-oxan-2-yl]propanoyl]amino]benzoic acid, CITRIC ACID, ... | | Authors: | Schaefer, M, Roehrig, S, Ackerstaff, J, Nunez, E.J, Gericke, K.M, Meier, K, Tersteegen, A, Stampfuss, J, Ellerbrock, P, Meibom, D, Lang, D, Heitmeier, S, Hillisch, A. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design and Preclinical Characterization Program toward Asundexian (BAY 2433334), an Oral Factor XIa Inhibitor for the Prevention and Treatment of Thromboembolic Disorders.
J.Med.Chem., 66, 2023
|
|
1RTI
 
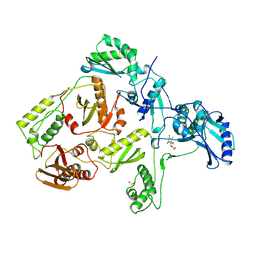 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | 1-(2-HYDROXYETHYLOXYMETHYL)-6-PHENYL THIOTHYMINE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-05-03 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
5IOI
 
 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
6W05
 
 | |
5I9Y
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with dasatinib | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.228 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
5IO9
 
 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
1RLC
 
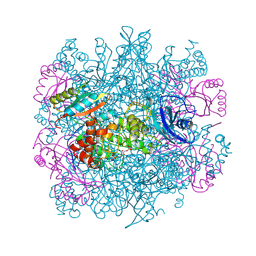 | | CRYSTAL STRUCTURE OF THE UNACTIVATED RIBULOSE 1, 5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE COMPLEXED WITH A TRANSITION STATE ANALOG, 2-CARBOXY-D-ARABINITOL 1,5-BISPHOSPHATE | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN) | | Authors: | Zhang, K.Y.J, Cascio, D, Eisenberg, D. | | Deposit date: | 1993-08-04 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the unactivated ribulose 1,5-bisphosphate carboxylase/oxygenase complexed with a transition state analog, 2-carboxy-D-arabinitol 1,5-bisphosphate.
Protein Sci., 3, 1994
|
|
5IP4
 
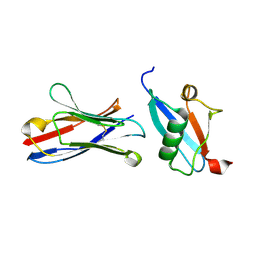 | | X-RAY STRUCTURE OF THE C-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin, XA4551 NANOBODY AGAINST C-DCX | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5LTP
 
 | |
6H2Y
 
 | | human Fab 1E6 bound to fHbp variant 3 from Neisseria meningitidis serogroup B | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Veggi, D, Bianchi, F, Cozzi, R, Malito, E, Bottomley, M.J. | | Deposit date: | 2018-07-17 | | Release date: | 2019-08-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Cocrystal structure of meningococcal factor H binding protein variant 3 reveals a new crossprotective epitope recognized by human mAb 1E6.
Faseb J., 33, 2019
|
|
3FPO
 
 | |
1JBD
 
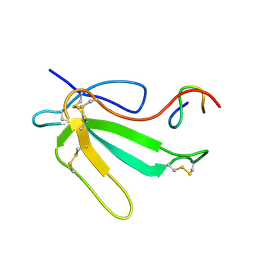 | | NMR Structure of the Complex Between alpha-bungarotoxin and a Mimotope of the Nicotinic Acetylcholine Receptor | | Descriptor: | LONG NEUROTOXIN 1, MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR | | Authors: | Scarselli, M, Spiga, O, Ciutti, A, Bracci, L, Lelli, B, Lozzi, L, Calamandrei, D, Bernini, A, Di Maro, D, Klein, S, Niccolai, N. | | Deposit date: | 2001-06-04 | | Release date: | 2001-06-27 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of alpha-bungarotoxin free and bound to a mimotope of the nicotinic acetylcholine receptor.
Biochemistry, 41, 2002
|
|
5EGM
 
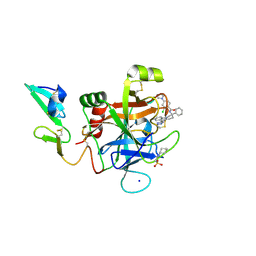 | | Development of a novel tricyclic class of potent and selective FIXa inhibitors | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-chloranyl-~{N}-[(7~{S})-2-methyl-7-phenyl-10-(1~{H}-1,2,3,4-tetrazol-5-yl)-8,9-dihydro-6~{H}-pyrido[1,2-a]indol-7-yl]-4-(1,2,4-triazol-4-yl)benzamide, Coagulation factor IX, ... | | Authors: | Meng, D, Andre, P, Bateman, T.J, Berger, R, Chen, Y, Desai, K, Dewnani, S, Ellsworth, K, Feng, D, Geissler, W.M, Guo, L, Hruza, A, Jian, T, Li, H, Parker, D.L, Reichert, P, Sherer, E.C, Smith, C.J, Sonatore, L.M, Tschirret-Guth, R, Wu, J, Xu, J, Zhang, T, Campeau, L, Orr, R, Poirier, M, McCabe-Dunn, j, Araki, K, Nishimura, T, Sakurada, I, Hirabayashi, T, Wood, H.B. | | Deposit date: | 2015-10-27 | | Release date: | 2015-11-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Development of a novel tricyclic class of potent and selective FIXa inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1JOJ
 
 | | CONCANAVALIN A-HEXAPEPTIDE COMPLEX | | Descriptor: | CALCIUM ION, Concanavalin-Br, HEXAPEPTIDE, ... | | Authors: | Jain, D, Kaur, K, Salunke, D.M. | | Deposit date: | 2001-07-30 | | Release date: | 2001-12-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Enhanced binding of a rationally designed peptide ligand of concanavalin a arises from improved geometrical complementarity.
Biochemistry, 40, 2001
|
|
6OOZ
 
 | | Asymmetric hTNF-alpha | | Descriptor: | (S)-{1-[(2,5-dimethylphenyl)methyl]-1H-benzimidazol-2-yl}(pyridin-4-yl)methanol, CHLORIDE ION, Tumor necrosis factor | | Authors: | Arakaki, T.L, Edwards, T.E, Fox III, D, Lecomte, F, Ceska, T. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Small molecules that inhibit TNF signalling by stabilising an asymmetric form of the trimer.
Nat Commun, 10, 2019
|
|
8CQ2
 
 | | Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, C16S, C20S, C870S, T1279C mutant | | Descriptor: | TcdA1 | | Authors: | Nganga, P.N, Roderer, D, Belyy, A, Prumbaum, D, Raunser, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-03-13 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Multistate kinetics of the syringe-like injection mechanism of Tc toxins.
Sci Adv, 11, 2025
|
|
8CPZ
 
 | | Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, K1179W mutant | | Descriptor: | TcdA1 | | Authors: | Nganga, P.N, Roderer, D, Belyy, A, Prumbaum, D, Raunser, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-03-13 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Multistate kinetics of the syringe-like injection mechanism of Tc toxins.
Sci Adv, 11, 2025
|
|
8CQ0
 
 | | Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, K567W K2008W mutant | | Descriptor: | TcdA1 | | Authors: | Nganga, P.N, Roderer, D, Belyy, A, Prumbaum, D, Raunser, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-03-13 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Multistate kinetics of the syringe-like injection mechanism of Tc toxins.
Sci Adv, 11, 2025
|
|
