3QDN
 
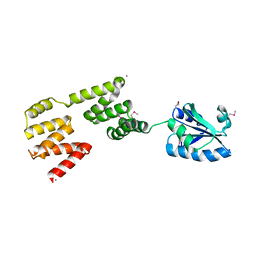 | | Putative thioredoxin protein from Salmonella typhimurium | | Descriptor: | CALCIUM ION, Putative thioredoxin protein | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-18 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Putative thioredoxin protein from Salmonella typhimurium.
To be Published
|
|
3QBO
 
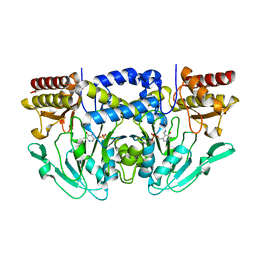 | | Crystal structure of phosphoserine aminotransferase from Yersinia pestis CO92 | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Phosphoserine aminotransferase | | Authors: | Nocek, B, Maltseva, N, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-13 | | Release date: | 2011-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structure of phosphoserine aminotransferase from Yersinia pestis CO92
TO BE PUBLISHED
|
|
3Q62
 
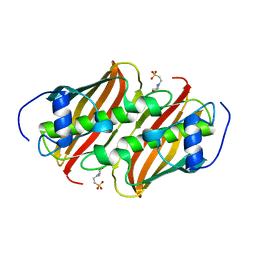 | | Crystal Structure of 3-hydroxydecanoyl-(acyl carrier protein) dehydratase from Yersinia pestis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase | | Authors: | Zhang, R, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-30 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of 3-hydroxydecanoyl-(acyl carrier protein) dehydratase from
Yersinia pestis
To be Published
|
|
3Q7I
 
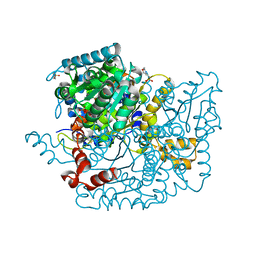 | | Glucose-6-phosphate isomerase from Francisella tularensis complexed with 6-phosphogluconic acid. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-PHOSPHOGLUCONIC ACID, CALCIUM ION, ... | | Authors: | Osipiuk, J, Maltseva, N, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-04 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Glucose-6-phosphate isomerase from Francisella tularensis.
To be Published
|
|
3QU1
 
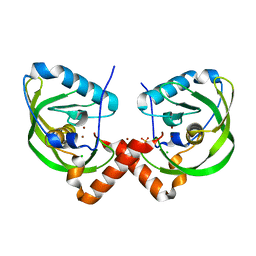 | | Peptide deformylase from Vibrio cholerae | | Descriptor: | CHLORIDE ION, Peptide deformylase 2, SULFATE ION, ... | | Authors: | Osipiuk, J, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-23 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptide deformylase from Vibrio cholerae.
To be Published
|
|
3PMM
 
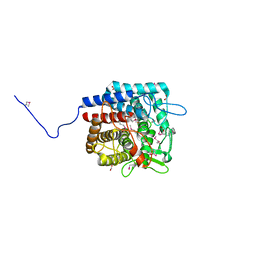 | | The crystal structure of a possible member of GH105 family from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 | | Descriptor: | FORMIC ACID, IMIDAZOLE, Putative cytoplasmic protein | | Authors: | Tan, K, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-17 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | The crystal structure of a possible member of GH105 family from Klebsiella pneumoniae subsp. pneumoniae MGH 78578
To be Published
|
|
3PN8
 
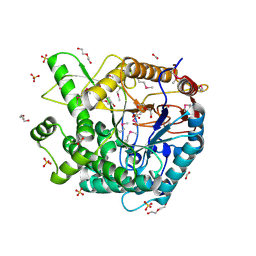 | | The crystal structure of 6-phospho-beta-glucosidase from Streptococcus mutans UA159 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, Putative phospho-beta-glucosidase, ... | | Authors: | Tan, K, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | The crystal structure of 6-phospho-beta-glucosidase from Streptococcus mutants UA159
To be Published
|
|
3PHA
 
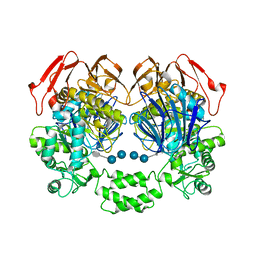 | | The crystal structure of the W169Y mutant of alpha-glucosidase (gh31 family) from Ruminococcus obeum atcc 29174 in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-glucosidase | | Authors: | Tan, K, Tesar, C, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-03 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.173 Å) | | Cite: | The crystal structure of the W169Y mutant of alpha-glucosidase (gh31 family) from Ruminococcus obeum atcc 29174 in complex with acarbose
To be Published
|
|
3PYW
 
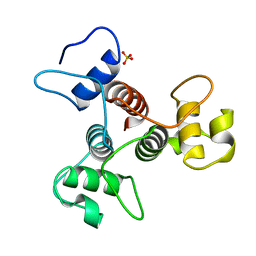 | | The structure of the SLH domain from B. anthracis surface array protein at 1.8A | | Descriptor: | S-layer protein sap, SULFATE ION | | Authors: | Zhang, R, Wilton, R, Kern, J, Joachimiak, A, Schneewind, O, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-12-13 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Surface Layer Homology (SLH) Domains from Bacillus anthracis Surface Array Protein.
J.Biol.Chem., 286, 2011
|
|
3POC
 
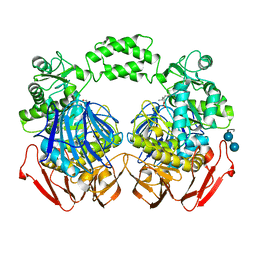 | | The crystal structure of the D307A mutant of alpha-Glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLYCEROL, ... | | Authors: | Tan, K, Tesar, C, Wilton, R, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-22 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the D307A mutant of alpha-Glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 in complex with acarbose
To be Published
|
|
3OOS
 
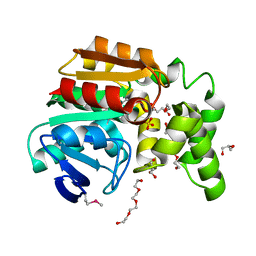 | | The structure of an alpha/beta fold family hydrolase from Bacillus anthracis str. Sterne | | Descriptor: | Alpha/beta hydrolase family protein, GLYCEROL, SULFATE ION, ... | | Authors: | Fan, Y, Tan, K, Bigelow, L, Hamilton, J, Li, H, Zhou, Y, Clancy, S, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-11-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of an alpha/beta fold family hydrolase from Bacillus anthracis str. Sterne
To be Published
|
|
3OWA
 
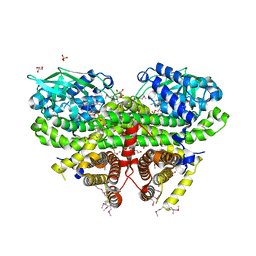 | | Crystal Structure of Acyl-CoA Dehydrogenase complexed with FAD from Bacillus anthracis | | Descriptor: | Acyl-CoA dehydrogenase, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kim, Y, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-17 | | Release date: | 2010-10-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of Acyl-CoA Dehydrogenase complexed with FAD from Bacillus anthracis
To be Published
|
|
3PEB
 
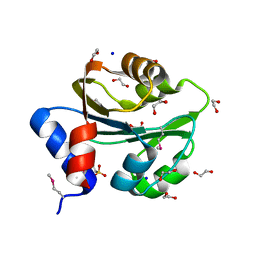 | | The Structure of a Creatine_N Superfamily domain of a dipeptidase from Streptococcus thermophilus. | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Dipeptidase, ... | | Authors: | Cuff, M.E, Mack, J.C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-10-25 | | Release date: | 2010-11-03 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Structure of a Creatine_N Superfamily domain of a dipeptidase from Streptococcus thermophilus.
TO BE PUBLISHED
|
|
3PEI
 
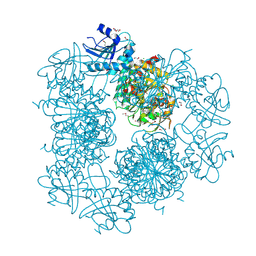 | | Crystal Structure of Cytosol Aminopeptidase from Francisella tularensis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cytosol aminopeptidase, ... | | Authors: | Maltseva, N, Kim, Y, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-26 | | Release date: | 2010-12-01 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Cytosol Aminopeptidase from
Francisella tularensis
To be Published
|
|
3QSG
 
 | |
3Q1H
 
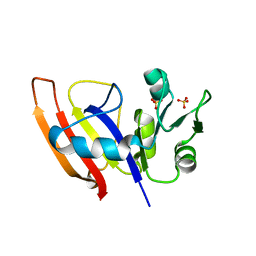 | | Crystal Structure of Dihydrofolate Reductase from Yersinia pestis | | Descriptor: | Dihydrofolate reductase, SULFATE ION | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-17 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Crystal Structure of Dihydrofolate Reductase from Yersinia pestis
To be Published
|
|
3Q12
 
 | | Pantoate-beta-alanine ligase from Yersinia pestis in complex with pantoate. | | Descriptor: | CHLORIDE ION, PANTOATE, Pantoate--beta-alanine ligase | | Authors: | Osipiuk, J, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-16 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Pantoate-beta-alanine ligase from Yersinia pestis.
To be Published
|
|
3Q1K
 
 | | The Crystal Structure of the D-alanyl-alanine Synthetase A from Salmonella enterica Typhimurium Complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine--D-alanine ligase A, FORMIC ACID, ... | | Authors: | Zhang, R, Maltseva, N, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-17 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | The Crystal Structure of the D-alanyl-alanine Synthetase A from Salmonella enterica Typhimurium Complexed with ADP
To be Published
|
|
3Q94
 
 | | The crystal structure of fructose 1,6-bisphosphate aldolase from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, ACETATE ION, Fructose-bisphosphate aldolase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-07 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | The crystal structure of fructose 1,6-bisphosphate aldolase from Bacillus anthracis str. 'Ames Ancestor'.
To be Published
|
|
3QZ6
 
 | |
3RNR
 
 | | Crystal Structure of Stage II Sporulation E Family Protein from Thermanaerovibrio acidaminovorans | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-22 | | Release date: | 2011-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Stage II Sporulation E Family Protein from Thermanaerovibrio acidaminovorans
To be Published
|
|
3RCN
 
 | | Crystal Structure of Beta-N-Acetylhexosaminidase from Arthrobacter aurescens | | Descriptor: | ACETIC ACID, Beta-N-acetylhexosaminidase, GLYCEROL, ... | | Authors: | Kim, Y, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-31 | | Release date: | 2011-06-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.514 Å) | | Cite: | Crystal Structure of Beta-N-Acetylhexosaminidase from Arthrobacter aurescens
To be Published
|
|
3RJ4
 
 | | Crystal Structure of 7-cyano-7-deazaguanine Reductase, QueF from Vibrio cholerae | | Descriptor: | 7-cyano-7-deazaguanine Reductase QueF, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-15 | | Release date: | 2011-08-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of 7-cyano-7-deazaguanine Reductase, QueF from Vibrio cholerae
To be Published
|
|
3RNS
 
 | |
3QJG
 
 | | Epidermin biosynthesis protein EpiD from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, Epidermin biosynthesis protein EpiD, FLAVIN MONONUCLEOTIDE | | Authors: | Osipiuk, J, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-28 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Epidermin biosynthesis protein EpiD from Staphylococcus aureus.
To be Published
|
|
