3NEG
 
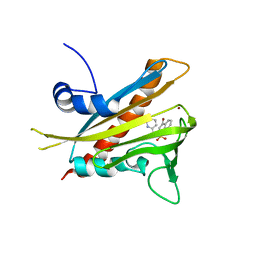 | | Pyrabactin-bound PYL1 structure in the open and close forms | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL1, BROMIDE ION | | Authors: | Yan, N. | | Deposit date: | 2010-06-08 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Functional mechanism of the ABA agonist pyrabactin
J.Biol.Chem., 285, 2010
|
|
5XOB
 
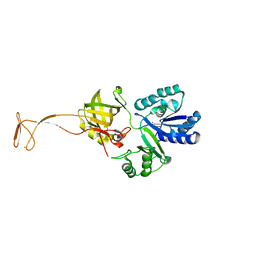 | | Crystal structure of apo TiaS (tRNAIle2 agmatidine synthetase) | | Descriptor: | MAGNESIUM ION, ZINC ION, tRNA(Ile2) 2-agmatinylcytidine synthetase TiaS | | Authors: | Dong, J. | | Deposit date: | 2017-05-27 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of tRNA-Modifying Enzyme TiaS and Motions of Its Substrate Binding Zinc Ribbon.
J. Mol. Biol., 430, 2018
|
|
3NEF
 
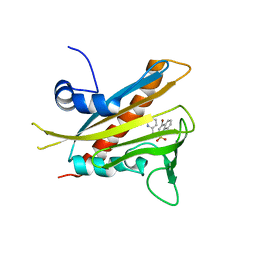 | | High-resolution pyrabactin-bound PYL1 structure | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL1 | | Authors: | Yan, N. | | Deposit date: | 2010-06-08 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional mechanism of the ABA agonist pyrabactin
J.Biol.Chem., 285, 2010
|
|
6LNU
 
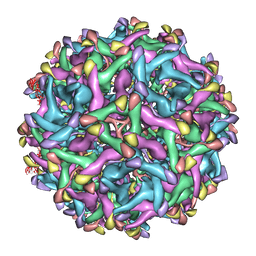 | | Cryo-EM structure of immature Zika virus | | Descriptor: | Genome polyprotein | | Authors: | Tan, T.Y, Fibriansah, G, Kostyuchenko, V.A, Ng, T.S, Lim, X.X, Lim, X.N, Shi, J, Morais, M.C, Corti, D, Lok, S.M. | | Deposit date: | 2020-01-02 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Capsid protein structure in Zika virus reveals the flavivirus assembly process.
Nat Commun, 11, 2020
|
|
6JQ5
 
 | | The structure of Hatchet Ribozyme | | Descriptor: | MAGNESIUM ION, RNA (82-MER) | | Authors: | Ren, A, Zheng, L. | | Deposit date: | 2019-03-29 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Hatchet ribozyme structure and implications for cleavage mechanism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5Z96
 
 | | Structure of the mouse TRPC4 ion channel | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Duan, J, Li, Z, Li, J, Zhang, J. | | Deposit date: | 2018-02-02 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the mouse TRPC4 ion channel.
Nat Commun, 9, 2018
|
|
4Q9Z
 
 | | Human Protein Kinase C Theta in Complex with Compound35 ((1R)-9-(AZETIDIN-3-YLAMINO)-1,8-DIMETHYL-3,5-DIHYDRO[1,2,4]TRIAZINO[3,4-C][1,4]BENZOXAZIN-2(1H)-ONE) | | Descriptor: | (1R)-9-(azetidin-3-ylamino)-1,8-dimethyl-3,5-dihydro[1,2,4]triazino[3,4-c][1,4]benzoxazin-2(1H)-one, HUMAN PROTEIN KINASE C THETA, SODIUM ION | | Authors: | Argiriadi, M.A, George, D.M. | | Deposit date: | 2014-05-02 | | Release date: | 2014-07-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Selective and Orally Bioavailable Protein Kinase C theta (PKC theta ) Inhibitors from a Fragment Hit.
J.Med.Chem., 58, 2015
|
|
6JG5
 
 | | Crystal structure of AimR | | Descriptor: | AimR transcriptional regulator | | Authors: | Guan, Z.Y, Pei, K, Zou, T.T. | | Deposit date: | 2019-02-13 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.221 Å) | | Cite: | Structural insights into DNA recognition by AimR of the arbitrium communication system in the SPbeta phage.
Cell Discov, 5, 2019
|
|
4QOY
 
 | |
6JG9
 
 | |
3O8X
 
 | | Recognition of Glycolipid Antigen by iNKT Cell TCR | | Descriptor: | (2S,3R)-3-HYDROXY-2-(TETRADECANOYLAMINO)OCTADECYL ALPHA-D-GALACTOPYRANOSIDURONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Li, Y. | | Deposit date: | 2010-08-03 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The V alpha 14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode.
J.Exp.Med., 207, 2010
|
|
3O9W
 
 | | Recognition of a Glycolipid Antigen by the iNKT Cell TCR | | Descriptor: | (2S)-3-(alpha-D-galactopyranosyloxy)-2-(hexadecanoyloxy)propyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Li, Y. | | Deposit date: | 2010-08-04 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The V alpha 14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode.
J.Exp.Med., 207, 2010
|
|
5CCP
 
 | | HISTIDINE 52 IS A CRITICAL RESIDUE FOR RAPID FORMATION OF CYTOCHROME C PEROXIDASE COMPOUND I | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Brown, K, Shaw, A, Miller, M.A, Kraut, J. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Histidine 52 is a critical residue for rapid formation of cytochrome c peroxidase compound I.
Biochemistry, 32, 1993
|
|
7X81
 
 | | The crystal structure of PloI4-C16M/D46A/I137V in complex with exo-2+2 adduct | | Descriptor: | (4S,4aS,6aR,8R,9R,11E,12aR,14aS,17E,18aR,18bR)-9-ethyl-4,8,19-trihydroxy-11,12a,13,18a-tetramethyl-2,3,4,4a,6a,7,8,9,10,12a,13,14,18a,18b-tetradecahydro-14a,17-(metheno)cyclobuta[b]naphtho[2,1-j][1]azacyclotetradecine-16,18(1H,15H)-dione, PloI4 | | Authors: | Li, M, Pan, L.F. | | Deposit date: | 2022-03-10 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | A cyclase that catalyses competing 2 + 2 and 4 + 2 cycloadditions.
Nat.Chem., 15, 2023
|
|
7X80
 
 | |
7X7Z
 
 | |
7X86
 
 | | The crystal structure of PloI4-F124L in complex with endo-4+2 adduct | | Descriptor: | (4S,4aS,6aR,8R,9R,10aS,13S,14aS,18aR,18bR,E)-9-ethyl-4,8,19-trihydroxy-10a,12,13,18a-tetramethyl-2,3,4,4a,6a,7,8,9,10,10a,13,14,18a,18b-tetradecahydro-14a,17-(metheno)benzo[b]naphtho[2,1-h][1]azacyclododecine-16,18(1H,15H)-dione, PloI4 | | Authors: | Li, M, Pan, L.F. | | Deposit date: | 2022-03-11 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | A cyclase that catalyses competing 2 + 2 and 4 + 2 cycloadditions.
Nat.Chem., 15, 2023
|
|
5FKF
 
 | |
5FKD
 
 | |
5FJC
 
 | |
5FK6
 
 | |
5FJ4
 
 | |
5FJ1
 
 | |
5FK1
 
 | |
5FKH
 
 | |
