7JQ5
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI8 | | Descriptor: | 3C-like proteinase, N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JQ0
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI3 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JQ2
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI5 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JQ1
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI4 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl]propan-2-yl}-L-phenylalaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
1HBV
 
 | |
5VPM
 
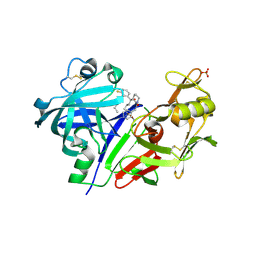 | |
5V8V
 
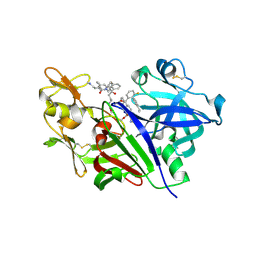 | | Crystal Structure of Human Renin in Complex with a biphenylpipderidinylcarbinol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, methyl [(4S)-4-(3'-ethyl-6-fluoro[1,1'-biphenyl]-2-yl)-4-hydroxy-4-{(3R)-1-[4-(methylamino)butanoyl]piperidin-3-yl}butyl]carbamate | | Authors: | Concha, N, Zhao, B. | | Deposit date: | 2017-03-22 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of renin inhibitors containing a simple aspartate binding moiety that imparts reduced P450 inhibition.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5VRP
 
 | |
7Y04
 
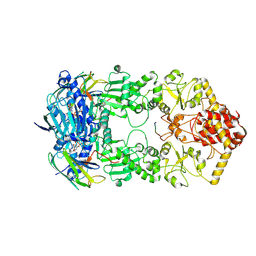 | | Hsp90-AhR-p23 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aryl hydrocarbon receptor, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Wen, Z.L, Zhai, Y.J, Zhu, Y, Sun, F. | | Deposit date: | 2022-06-03 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the cytosolic AhR complex.
Structure, 31, 2023
|
|
8H77
 
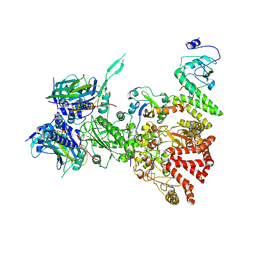 | | Hsp90-AhR-p23-XAP2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, Aryl hydrocarbon receptor, ... | | Authors: | Wen, Z.L, Zhai, Y.J, Zhu, Y, Sun, F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the cytosolic AhR complex.
Structure, 31, 2023
|
|
7U9Y
 
 | |
3N75
 
 | | X-ray Crystal Structure of the Escherichia coli Inducible Lysine Decarboxylase LdcI | | Descriptor: | GLYCEROL, GUANOSINE-5',3'-TETRAPHOSPHATE, HEXAETHYLENE GLYCOL, ... | | Authors: | Kanjee, U, Alexopoulos, E, Pai, E.F, Houry, W.A. | | Deposit date: | 2010-05-26 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase.
Embo J., 30, 2011
|
|
3NBX
 
 | |
6CV7
 
 | |
6DY7
 
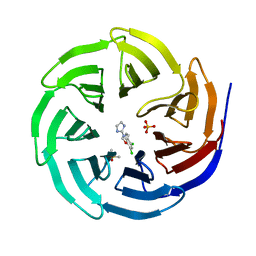 | | WDR5 in complex with a WIN site inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, N-[3-(2,4-dichlorophenoxy)propyl]-1H-imidazol-2-amine, SULFATE ION, ... | | Authors: | Phan, J, Wang, F, Fesik, S.W. | | Deposit date: | 2018-07-01 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Displacement of WDR5 from Chromatin by a WIN Site Inhibitor with Picomolar Affinity.
Cell Rep, 26, 2019
|
|
6DYA
 
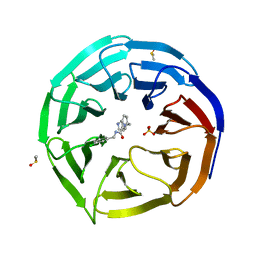 | | WDR5 in complex with a WIN site inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3,5-dichlorophenyl)methyl]-3-[(1H-imidazol-1-yl)methyl]benzamide, SULFATE ION, ... | | Authors: | Phan, J, Wang, F, Fesik, S.W. | | Deposit date: | 2018-07-01 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Displacement of WDR5 from Chromatin by a WIN Site Inhibitor with Picomolar Affinity.
Cell Rep, 26, 2019
|
|
6E1Y
 
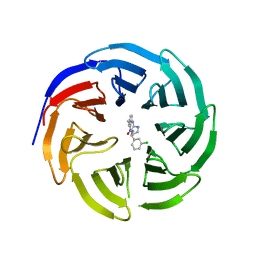 | |
6E23
 
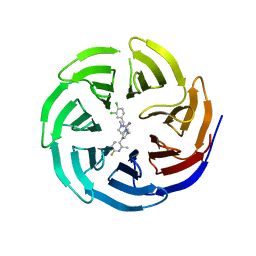 | |
6E22
 
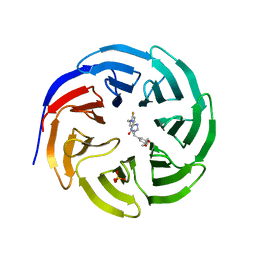 | |
6E1Z
 
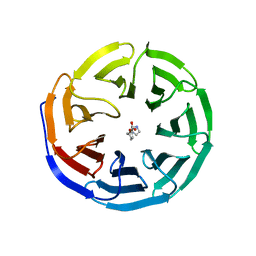 | |
1SN4
 
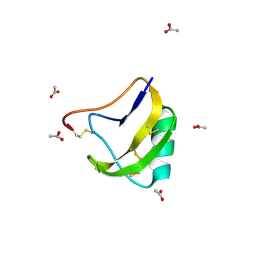 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M4 | | Descriptor: | ACETATE ION, PROTEIN (NEUROTOXIN BMK M4) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-11 | | Release date: | 1999-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
1SNB
 
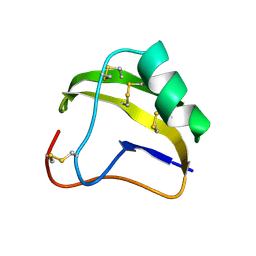 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M8 | | Descriptor: | NEUROTOXIN BMK M8 | | Authors: | Wang, D.C, Zeng, Z.H, Li, H.M. | | Deposit date: | 1997-03-12 | | Release date: | 1997-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an acidic neurotoxin from scorpion Buthus martensii Karsch at 1.85 A resolution.
J.Mol.Biol., 261, 1996
|
|
1SN1
 
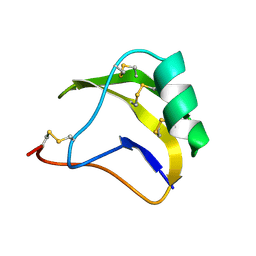 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M1 | | Descriptor: | PROTEIN (NEUROTOXIN BMK M1) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
5FFG
 
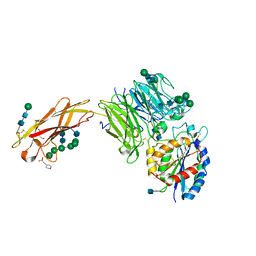 | | Crystal structure of integrin alpha V beta 6 head | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dong, X, Springer, T.A. | | Deposit date: | 2015-12-18 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Force interacts with macromolecular structure in activation of TGF-beta.
Nature, 542, 2017
|
|
6MCW
 
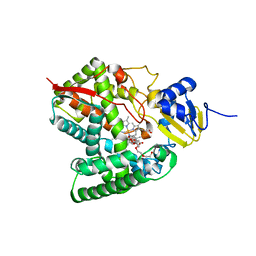 | | Crystal structure of the P450 domain of the CYP51-ferredoxin fusion protein from Methylococcus capsulatus, complex with the detergent Anapoe-X-114 | | Descriptor: | 23-[4-(2,4,4-trimethylpentan-2-yl)phenoxy]-3,6,9,12,15,18,21-heptaoxatricosan-1-ol, Cytochrome P450 51, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hargrove, T, Wawrzak, Z, Lamb, D.C, Lepesheva, G.I. | | Deposit date: | 2018-09-02 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Concerning P450 evolution: Structural Analyses Support Bacterial Origin of Sterol 14 alpha-Demethylases.
Mol.Biol.Evol., 2020
|
|
