5JQM
 
 | | Crystal Structure of Phosphatidic acid Transporter Ups1/Mdm35 Void of Bound Phospholipid from Saccharomyces Cerevisiae at 1.5 Angstroms Resolution | | Descriptor: | Protein UPS1, mitochondrial,Mitochondrial distribution and morphology protein 35 | | Authors: | Lu, J, Chan, K.C, Zhai, Y, Fan, J, Sun, F. | | Deposit date: | 2016-05-05 | | Release date: | 2017-07-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular mechanism of mitochondrial phosphatidate transfer by Ups1
Commun Biol, 2020
|
|
6EUI
 
 | | The GH43, Beta 1,3 Galactosidase, BT3683 with galactose | | Descriptor: | Beta-glucanase, CALCIUM ION, beta-D-galactopyranose | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
6EUJ
 
 | | The GH43, Beta 1,3 Galactosidase, BT0265 | | Descriptor: | Beta-glucanase | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
6EUF
 
 | | The GH43, Beta 1,3 Galactosidase, BT0265 | | Descriptor: | Beta-glucanase, alpha-L-arabinofuranose-(1-3)-[alpha-L-arabinofuranose-(1-4)][beta-D-glucopyranuronic acid-(1-6)]beta-D-galactopyranose-(1-6)-beta-D-galactopyranose, alpha-L-rhamnopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-6)-[alpha-L-arabinofuranose-(1-3)][alpha-L-arabinofuranose-(1-4)]beta-D-galactopyranose-(1-6)-beta-D-galactopyranose | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
6EUH
 
 | | The GH43, Beta 1,3 Galactosidase, BT3683 with galactodeoxynojirimycin | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, Beta-glucanase, CALCIUM ION | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
6EUG
 
 | |
7E1Q
 
 | | Crystal structure of dehydrogenase/isomerase FabX from Helicobacter pylori | | Descriptor: | 2-nitropropane dioxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Helicobacter pylori FabX contains a [4Fe-4S] cluster essential for unsaturated fatty acid synthesis.
Nat Commun, 12, 2021
|
|
7E1R
 
 | | Crystal structure of Dehydrogenase/isomerase FabX from Helicobacter pylori in complex with holo-ACP | | Descriptor: | 2-nitropropane dioxygenase, Acyl carrier protein,Acyl carrier protein, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | Helicobacter pylori FabX contains a [4Fe-4S] cluster essential for unsaturated fatty acid synthesis.
Nat Commun, 12, 2021
|
|
7E1S
 
 | | Crystal structure of dehydrogenase/isomerase FabX from Helicobacter pylori in complex with octanoyl-ACP | | Descriptor: | 2-nitropropane dioxygenase, Acyl carrier protein,Acyl carrier protein, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Helicobacter pylori FabX contains a [4Fe-4S] cluster essential for unsaturated fatty acid synthesis.
Nat Commun, 12, 2021
|
|
8YA1
 
 | | HEN EGG WHITE LYSOZYME | | Descriptor: | Lysozyme C | | Authors: | Zhang, C.Y, Xu, Q, Wang, W.W, Zhou, H. | | Deposit date: | 2024-02-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic data collection using a multilayer monochromator on an undulator beamline at SSRF
To Be Published
|
|
8YA5
 
 | |
8YA4
 
 | |
8Y9W
 
 | |
8Y9V
 
 | | ZIKV NS2B/NS3 protease | | Descriptor: | DAR-LYS-ORN-ARG, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Zhang, C.Y, Xu, Q, Wang, W.W, Zhou, H, Wang, Q.S. | | Deposit date: | 2024-02-07 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic data collection using a multilayer monochromator on an undulator beamline at SSRF
To Be Published
|
|
6EON
 
 | | Galactanase BT0290 | | Descriptor: | Beta-galactosidase, CALCIUM ION, alpha-D-galactopyranose | | Authors: | Basle, A, Munoz, J, Gilbert, H. | | Deposit date: | 2017-10-10 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
6KYL
 
 | | Crystal Structure of Phosphatidic acid Transporter Ups1/Mdm35 in Complex with (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, Mitochondrial distribution and morphology protein 35, Protein UPS1, ... | | Authors: | Lu, J, Chan, K.C, Zhai, Y, Fan, J, Sun, F. | | Deposit date: | 2019-09-19 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Molecular mechanism of mitochondrial phosphatidate transfer by Ups1.
Commun Biol, 3, 2020
|
|
6ADQ
 
 | | Respiratory Complex CIII2CIV2SOD2 from Mycobacterium smegmatis | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Gong, H.R, Xu, A, Gao, R.G, Ji, W.X, Wang, S.H, Wang, Q, Li, J, Rao, Z.H. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An electron transfer path connects subunits of a mycobacterial respiratory supercomplex.
Science, 362, 2018
|
|
6LUM
 
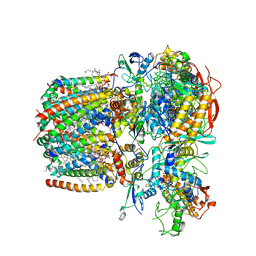 | | Structure of Mycobacterium smegmatis succinate dehydrogenase 2 | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ... | | Authors: | Gao, Y, Gong, H, Zhou, X, Xiao, Y, Wang, W, Ji, W, Wang, Q, Rao, Z. | | Deposit date: | 2020-01-29 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of trimeric Mycobacterium smegmatis succinate dehydrogenase with a membrane-anchor SdhF.
Nat Commun, 11, 2020
|
|
4FWJ
 
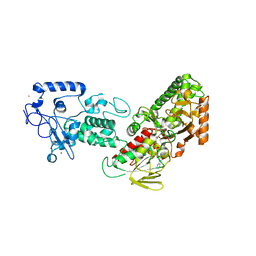 | | Native structure of LSD2/AOF1/KDM1b in spacegroup of I222 at 2.9A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, PHOSPHATE ION, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
4FWE
 
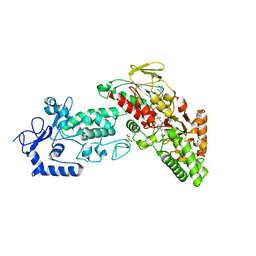 | | Native structure of LSD2 /AOF1/KDM1b in spacegroup of C2221 at 2.13A | | Descriptor: | CITRATE ANION, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
4FWF
 
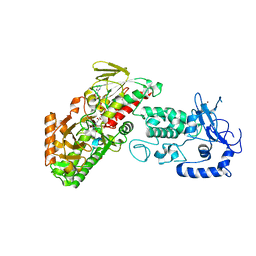 | | Complex structure of LSD2/AOF1/KDM1b with H3K4 mimic | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.1, Lysine-specific histone demethylase 1B, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
5W1L
 
 | |
5W1J
 
 | | Echinococcus granulosus thioredoxin glutathione reductas (egTGR) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin glutathione reductase | | Authors: | Gao, W, Wang, Y, Dai, S. | | Deposit date: | 2017-06-03 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Enzymatic and Structural Basis for Inhibition of Echinococcus granulosus Thioredoxin Glutathione Reductase by Gold(I).
Antioxid. Redox Signal., 27, 2017
|
|
6CHZ
 
 | | Estrogen Receptor Alpha Y537S bound to antagonist H3B-9224. | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-{4-[(1E)-1-(1H-indazol-5-yl)-2-phenylbut-1-en-1-yl]phenoxy}ethyl)amino]-N,N-dimethylbutanamide, Estrogen receptor | | Authors: | Larsen, N.A. | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of Selective Estrogen Receptor Covalent Antagonists for the Treatment of ER alphaWTand ER alphaMUTBreast Cancer.
Cancer Discov, 8, 2018
|
|
6CHW
 
 | | Estrogen Receptor Alpha Y537S covalently bound to antagonist H3B-5942. | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-{4-[(1E)-1-(1H-indazol-5-yl)-2-phenylbut-1-en-1-yl]phenoxy}ethyl)amino]-N,N-dimethylbutanamide, DIMETHYL SULFOXIDE, ... | | Authors: | Larsen, N.A. | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Discovery of Selective Estrogen Receptor Covalent Antagonists for the Treatment of ER alphaWTand ER alphaMUTBreast Cancer.
Cancer Discov, 8, 2018
|
|
