8WHI
 
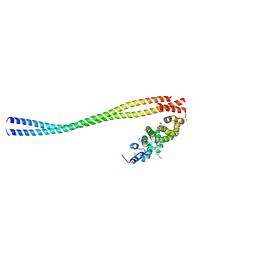 | |
8WHK
 
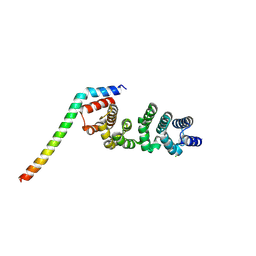 | | Crystal structure of CLASP2 in complex with LL5beta | | Descriptor: | CHLORIDE ION, CLIP-associating protein 2, Pleckstrin homology-like domain family B member 2 | | Authors: | Jia, X, Wei, Z. | | Deposit date: | 2023-09-23 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | CLASP-mediated competitive binding in protein condensates directs microtubule growth.
Nat Commun, 15, 2024
|
|
8WHJ
 
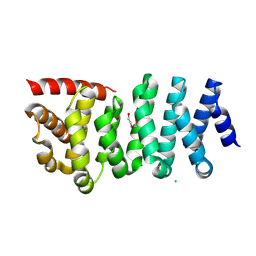 | | Crystal structure of CLASP2 TOG4 fused with LL5beta | | Descriptor: | CHLORIDE ION, CLIP-associating protein 2,Pleckstrin homology-like domain family B member 2, DI(HYDROXYETHYL)ETHER | | Authors: | Jia, X, Wei, Z. | | Deposit date: | 2023-09-23 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | CLASP-mediated competitive binding in protein condensates directs microtubule growth.
Nat Commun, 15, 2024
|
|
8WHL
 
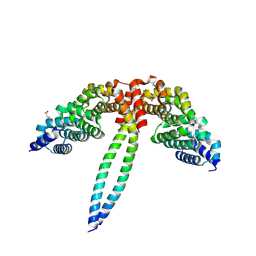 | | Crystal structure of CLASP2 in complex with CENP-E | | Descriptor: | ACETATE ION, CLIP-associating protein 2, Centromere-associated protein E, ... | | Authors: | Jia, X, Wei, Z. | | Deposit date: | 2023-09-23 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | CLASP-mediated competitive binding in protein condensates directs microtubule growth.
Nat Commun, 15, 2024
|
|
8WHH
 
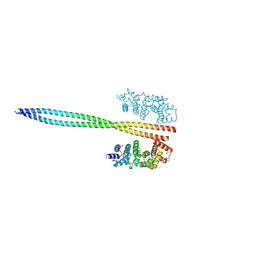 | |
8WHM
 
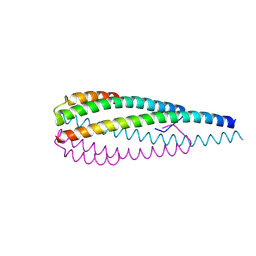 | | Crystal structure of the ELKS2/LL5beta complex | | Descriptor: | ERC protein 2, Pleckstrin homology-like domain family B member 2 | | Authors: | Jia, X, Wei, Z. | | Deposit date: | 2023-09-23 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CLASP-mediated competitive binding in protein condensates directs microtubule growth.
Nat Commun, 15, 2024
|
|
5Y45
 
 | |
7EY5
 
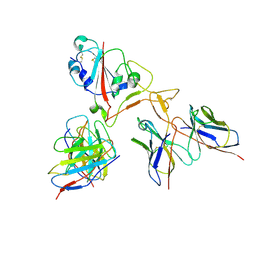 | |
7EZV
 
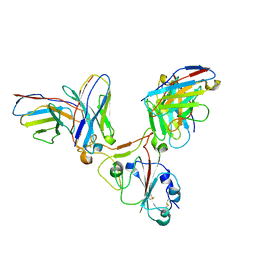 | |
7EY4
 
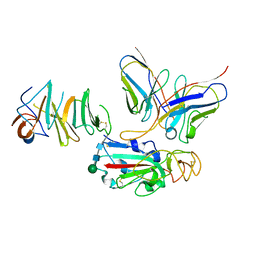 | | Local CryoEM of the SARS-CoV-2 S6PV2 in complex with BD-667 | | Descriptor: | BD-667 H, BD-667 L, Spike glycoprotein, ... | | Authors: | Liu, P.L. | | Deposit date: | 2021-05-29 | | Release date: | 2021-09-08 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structures of SARS-CoV-2 B.1.351 neutralizing antibodies provide insights into cocktail design against concerning variants.
Cell Res., 31, 2021
|
|
7WP6
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 36H6 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-01 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7WP8
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK1628x in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B4 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
5Y46
 
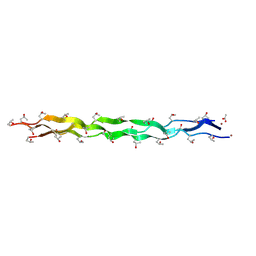 | |
7WB5
 
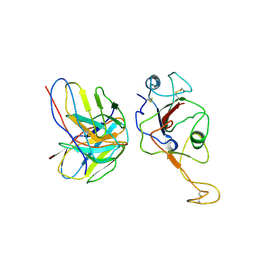 | | local structure of hu33 and spike | | Descriptor: | Surface glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, hu33 heavy chain, ... | | Authors: | Pulan, L. | | Deposit date: | 2021-12-15 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A non-ACE2-blocking neutralizing antibody against Omicron-included SARS-CoV-2 variants.
Signal Transduct Target Ther, 7, 2022
|
|
7WBH
 
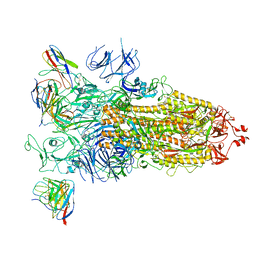 | | overall structure of hu33 and spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Pulan, L. | | Deposit date: | 2021-12-16 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A non-ACE2-blocking neutralizing antibody against Omicron-included SARS-CoV-2 variants.
Signal Transduct Target Ther, 7, 2022
|
|
8W41
 
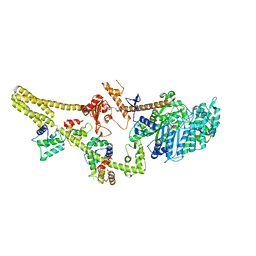 | | Cryo-EM structure of Myosin VI in the autoinhibited state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Calmodulin-1, ... | | Authors: | Niu, F, Wei, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Autoinhibition and activation of myosin VI revealed by its cryo-EM structure.
Nat Commun, 15, 2024
|
|
6KWY
 
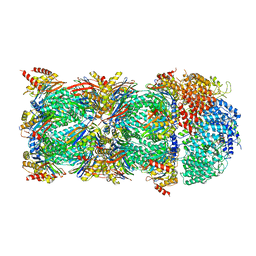 | | human PA200-20S complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Proteasome activator complex subunit 4, Proteasome subunit alpha type-1, ... | | Authors: | Ouyang, S, Hongxin, G. | | Deposit date: | 2019-09-09 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Cryo-EM structures of the human PA200 and PA200-20S complex reveal regulation of proteasome gate opening and two PA200 apertures.
Plos Biol., 18, 2020
|
|
6A6H
 
 | |
7W0S
 
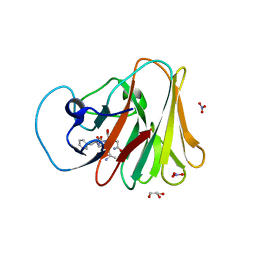 | | TRIM7 in complex with C-terminal peptide of 2C | | Descriptor: | DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase TRIM7, GLYCEROL, ... | | Authors: | Zhang, H, Liang, X, Li, X.Z. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A C-terminal glutamine recognition mechanism revealed by E3 ligase TRIM7 structures.
Nat.Chem.Biol., 18, 2022
|
|
7W0Q
 
 | | TRIM7 in complex with C-terminal peptide of 2C | | Descriptor: | E3 ubiquitin-protein ligase TRIM7, peptide | | Authors: | Zhang, H, Liang, X, Li, X.Z. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A C-terminal glutamine recognition mechanism revealed by E3 ligase TRIM7 structures.
Nat.Chem.Biol., 18, 2022
|
|
7W0T
 
 | | TRIM7 in complex with C-terminal peptide of 2C | | Descriptor: | E3 ubiquitin-protein ligase TRIM7, peptide | | Authors: | Zhang, H, Liang, X, Li, X.Z. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A C-terminal glutamine recognition mechanism revealed by E3 ligase TRIM7 structures.
Nat.Chem.Biol., 18, 2022
|
|
