1B26
 
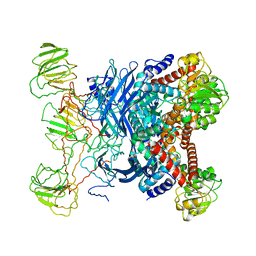 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE | | Authors: | Knapp, S, Devos, W.M, Rice, D, Ladenstein, R. | | Deposit date: | 1998-12-04 | | Release date: | 1999-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of glutamate dehydrogenase from the hyperthermophilic eubacterium Thermotoga maritima at 3.0 A resolution.
J.Mol.Biol., 267, 1997
|
|
1CM0
 
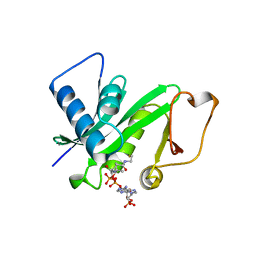 | | CRYSTAL STRUCTURE OF THE PCAF/COENZYME-A COMPLEX | | Descriptor: | COENZYME A, P300/CBP ASSOCIATING FACTOR | | Authors: | Clements, A, Rojas, J.R, Trievel, R.C, Wang, L, Berger, S.L, Marmorstein, R. | | Deposit date: | 1999-05-12 | | Release date: | 1999-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A.
EMBO J., 18, 1999
|
|
1QSN
 
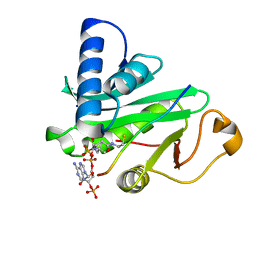 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 WITH BOUND COENZYME A AND HISTONE H3 PEPTIDE | | Descriptor: | COENZYME A, HISTONE H3, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-22 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
1QST
 
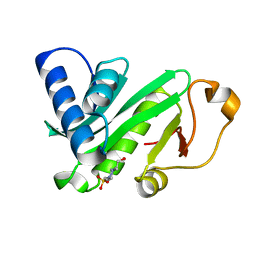 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
4F2U
 
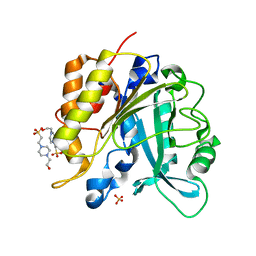 | | Structure of the N254Y/H258Y double mutant of the Phosphatidylinositol-Specific Phospholipase C from S.aureus | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION | | Authors: | Cheng, J, Goldstein, R, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-05-08 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Competition between Anion Binding and Dimerization Modulates Staphylococcus aureus Phosphatidylinositol-specific Phospholipase C Enzymatic Activity.
J.Biol.Chem., 287, 2012
|
|
4E26
 
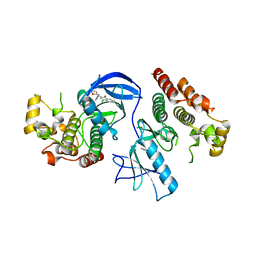 | | BRAF in complex with an organic inhibitor 7898734 | | Descriptor: | 5-chloro-7-[(R)-furan-2-yl(pyridin-2-ylamino)methyl]quinolin-8-ol, Serine/threonine-protein kinase B-raf | | Authors: | Qin, J, Xie, P, Ventocilla, C, Zhou, G, Vultur, A, Chen, Q, Herlyn, M, Winkler, J, Marmorstein, R. | | Deposit date: | 2012-03-07 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification of a Novel Family of BRAF(V600E) Inhibitors.
J.Med.Chem., 55, 2012
|
|
1QSR
 
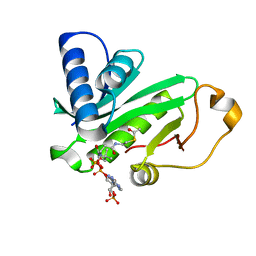 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 WITH BOUND ACETYL-COENZYME A | | Descriptor: | ACETYL COENZYME *A, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
2JFB
 
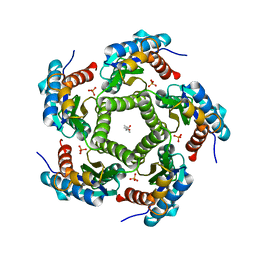 | | 3D Structure of Lumazine Synthase from Candida albicans | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, PHOSPHATE ION | | Authors: | Morgunova, E, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lumazine Synthase from Candida Albicans as an Anti- Fungal Target Enzyme: Structural and Biochemical Basis for Drug Design.
J.Biol.Chem., 282, 2007
|
|
1PKV
 
 | | The N-terminal domain of riboflavin synthase in complex with riboflavin | | Descriptor: | RIBOFLAVIN, Riboflavin synthase alpha chain | | Authors: | Meining, W, Eberhardt, S, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of the N-terminal domain of riboflavin synthase in complex with riboflavin at 2.6A resolution.
J.Mol.Biol., 331, 2003
|
|
1Q2C
 
 | |
4EQC
 
 | | Crystal structure of PAK1 kinase domain in complex with FRAX597 inhibitor | | Descriptor: | 6-[2-chloro-4-(1,3-thiazol-5-yl)phenyl]-8-ethyl-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, CHLORIDE ION, Serine/threonine-protein kinase PAK 1 | | Authors: | Maksimoska, J, Marmorstein, R. | | Deposit date: | 2012-04-18 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | FRAX597, a Small Molecule Inhibitor of the p21-activated Kinases, Inhibits Tumorigenesis of Neurofibromatosis Type 2 (NF2)-associated Schwannomas.
J.Biol.Chem., 288, 2013
|
|
1S5P
 
 | | Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Eschericia coli. | | Descriptor: | HISTONE H4 (RESIDUES 12-19), NAD-dependent deacetylase, ZINC ION | | Authors: | Zhao, K, Chai, X, Marmorstein, R. | | Deposit date: | 2004-01-21 | | Release date: | 2004-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure and Substrate Binding Properties of cobB, a Sir2 Homolog Protein Deacetylase from Eschericia coli.
J.Mol.Biol., 337, 2004
|
|
4GS4
 
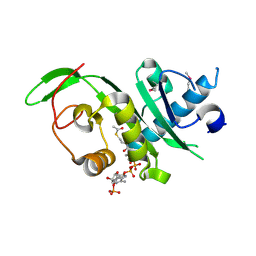 | | Structure of the alpha-tubulin acetyltransferase, alpha-TAT1 | | Descriptor: | ACETYL COENZYME *A, Alpha-tubulin N-acetyltransferase | | Authors: | Friedmann, D.R, Fan, J, Marmorstein, R. | | Deposit date: | 2012-08-27 | | Release date: | 2012-10-17 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Structure of the alpha-tubulin acetyltransferase, alpha TAT1, and implications for tubulin-specific acetylation.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1PU9
 
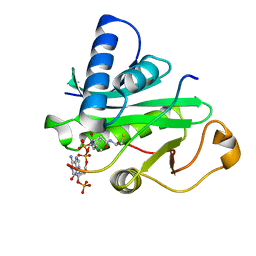 | | Crystal Structure of Tetrahymena GCN5 with Bound Coenzyme A and a 19-residue Histone H3 Peptide | | Descriptor: | COENZYME A, HAT A1, Histone H3 | | Authors: | Clements, A, Poux, A.N, Lo, W.S, Pillus, L, Berger, S.L, Marmorstein, R. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for histone and phospho-histone binding by the GCN5 histone acetyltransferase
Mol.Cell, 12, 2003
|
|
1Q2D
 
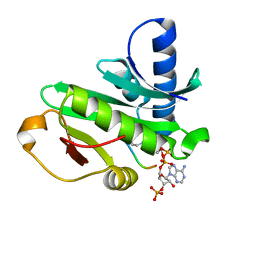 | |
3RQ3
 
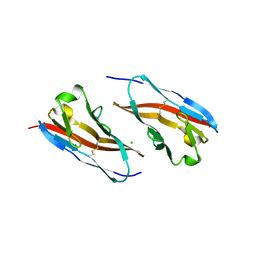 | | Structure of T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT) in hexagonal crystal form | | Descriptor: | CHLORIDE ION, T cell immunoreceptor with Ig and ITIM domains | | Authors: | Ramagopal, U.A, Rubinstein, R, Guo, H, Samanta, D, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2011-04-27 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT) in hexagonal crystal form
To be published
|
|
3Q4C
 
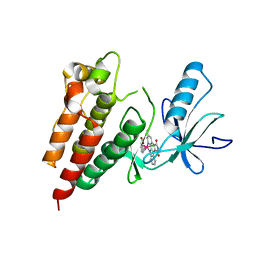 | | Crystal Structure of Wild Type BRAF kinase domain in complex with organometallic inhibitor CNS292 | | Descriptor: | Serine/threonine-protein kinase B-raf, [(1,2,3,4,5,6-eta)-(1S,2R,3R,4R,5S,6S)-1-carboxycyclohexane-1,2,3,4,5,6-hexayl](chloro)(3-methyl-5,7-dioxo-6,7-dihydro-5H-pyrido[2,3-a]pyrrolo[3,4-c]carbazol-12-ide-kappa~2~N~1~,N~12~)ruthenium(1+) | | Authors: | Xie, P, Streu, C, Qin, J, Pregman, H, Pagano, N, Meggers, E, Marmorstein, R. | | Deposit date: | 2010-12-23 | | Release date: | 2011-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of BRAF in complex with an organoruthenium inhibitor reveals a mechanism for inhibition of an active form of BRAF kinase.
Biochemistry, 48, 2009
|
|
4F2T
 
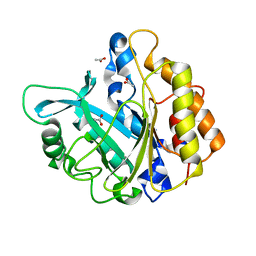 | | Modulation of S.aureus Phosphatidylinositol-Specific Phospholipase C Membrane Binding. | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | Authors: | Cheng, J, Goldstein, R, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-05-08 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Competition between Anion Binding and Dimerization Modulates Staphylococcus aureus Phosphatidylinositol-specific Phospholipase C Enzymatic Activity.
J.Biol.Chem., 287, 2012
|
|
4F2B
 
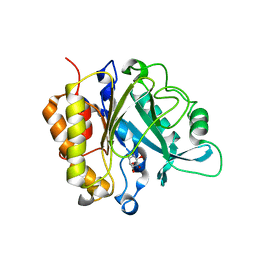 | | Modulation of S.Aureus Phosphatidylinositol-Specific Phospholipase C Membrane Binding | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase | | Authors: | Cheng, J, Goldstein, R, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-05-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Competition between Anion Binding and Dimerization Modulates Staphylococcus aureus Phosphatidylinositol-specific Phospholipase C Enzymatic Activity.
J.Biol.Chem., 287, 2012
|
|
1Q14
 
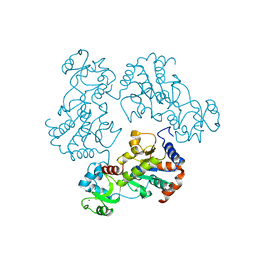 | | Structure and autoregulation of the yeast Hst2 homolog of Sir2 | | Descriptor: | CHLORIDE ION, HST2 protein, ZINC ION | | Authors: | Zhao, K, Chai, X, Clements, A, Marmorstein, R. | | Deposit date: | 2003-07-18 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and autoregulation of the Yeast Hst2 homolog of Sir2
Nat.Struct.Biol., 10, 2003
|
|
1PUA
 
 | | Crystal Structure of Tetrahymena GCN5 with Bound Coenzyme A and a Phosphorylated, 19-residue Histone H3 peptide | | Descriptor: | COENZYME A, HAT A1, Histone H3 | | Authors: | Clements, A, Poux, A.N, Lo, W.S, Pillus, L, Berger, S.L, Marmorstein, R. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for histone and phospho-histone binding by the GCN5 histone acetyltransferase
Mol.Cell, 12, 2003
|
|
3QM0
 
 | | Crystal structure of RTT109-AC-CoA complex | | Descriptor: | ACETYL COENZYME *A, Histone acetyltransferase RTT109, MERCURY (II) ION | | Authors: | Tang, Y, Marmorstein, R. | | Deposit date: | 2011-02-03 | | Release date: | 2011-02-16 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2P4W
 
 | | Crystal structure of heat shock regulator from Pyrococcus furiosus | | Descriptor: | SULFATE ION, Transcriptional regulatory protein arsR family | | Authors: | Liu, W, Vierke, G, Panjikar, S, Thomm, M, Ladenstein, R. | | Deposit date: | 2007-03-13 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Archaeal Heat Shock Regulator from Pyrococcus furiosus: A Molecular Chimera Representing Eukaryal and Bacterial Features.
J.Mol.Biol., 369, 2007
|
|
3HI8
 
 | | Crystal structure of proliferating cell nuclear antigen (PCNA) from Haloferax volcanii | | Descriptor: | Proliferating cell nuclear antigen PcnA | | Authors: | Morgunova, E, Gray, F.C, MacNeill, S.A, Ladenstein, R. | | Deposit date: | 2009-05-19 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural insights into the adaptation of proliferating cell nuclear antigen (PCNA) from Haloferax volcanii to a high-salt environment.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3FXZ
 
 | |
