3JSX
 
 | | X-ray Crystal structure of NAD(P)H: Quinone Oxidoreductase-1 (NQO1) bound to the coumarin-based inhibitor AS1 | | Descriptor: | 4-hydroxy-6,7-dimethyl-3-(naphthalen-1-ylmethyl)-2H-chromen-2-one, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Dunstan, M.S, Levy, C, Leys, D. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthesis and biological evaluation of coumarin-based inhibitors of NAD(P)H: quinone oxidoreductase-1 (NQO1).
J.Med.Chem., 52, 2009
|
|
4LX6
 
 | |
4LX5
 
 | | X-ray crystal structure of the M6" riboswitch aptamer bound to pyrimido[4,5-d]pyrimidine-2,4-diamine (PPDA) | | Descriptor: | MAGNESIUM ION, Mutated adenine riboswitch aptamer, pyrimido[4,5-d]pyrimidine-2,4-diamine | | Authors: | Dunstan, M.S, Leys, D. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Modular riboswitch toolsets for synthetic genetic control in diverse bacterial species.
J.Am.Chem.Soc., 136, 2014
|
|
4KVR
 
 | | Crystal Structure of Prochlorococcus marinus aldehyde-deformylating oxygenase (mutant V41Y) | | Descriptor: | Aldehyde decarbonylase, FE (III) ION, HEXANOIC ACID | | Authors: | Levy, C.W, Khara, B, Menon, N, Mansell, D, Das, D, Marsh, E.N.G, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase.
Chembiochem, 14, 2013
|
|
4KVS
 
 | | Crystal Structure of Prochlorococcus marinus aldehyde-deformylating oxygenase (mutant A134F) | | Descriptor: | Aldehyde decarbonylase, FE (III) ION, HEXANOIC ACID | | Authors: | Levy, C.W, Khara, B, Menon, N, Mansell, D, Das, D, Marsh, E.N.G, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase.
Chembiochem, 14, 2013
|
|
3EKD
 
 | |
3F03
 
 | |
3EKF
 
 | |
3GSI
 
 | | Crystal structure of D552A dimethylglycine oxidase mutant of Arthrobacter globiformis in complex with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Tralau, T, Lafite, P, Levy, C, Combe, J.P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An internal reaction chamber in dimethylglycine oxidase provides efficient protection from exposure to toxic formaldehyde.
J.Biol.Chem., 284, 2009
|
|
3GX9
 
 | | Structure of morphinone reductase N189A mutant in complex with tetrahydroNAD | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN MONONUCLEOTIDE, Morphinone reductase | | Authors: | Lafite, P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Parallel Pathways and Free-Energy Landscapes for Enzymatic Hydride Transfer Probed by Hydrostatic Pressure
Chembiochem, 10, 2009
|
|
3DG9
 
 | | Crystal Structure of Malonate Decarboxylase from Bordatella bronchiseptica | | Descriptor: | Arylmalonate decarboxylase, PHOSPHATE ION | | Authors: | Okrasa, K, Levy, C, Baudendistel, N, Leys, D, Micklefield, J. | | Deposit date: | 2008-06-13 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Mechanism of an Unusual Malonate Decarboxylase and Related Racemases.
Chemistry, 14, 2008
|
|
4DQL
 
 | |
4EPQ
 
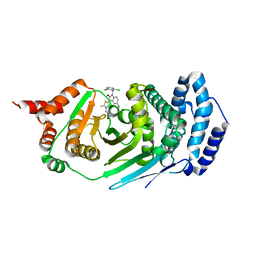 | |
4O0Q
 
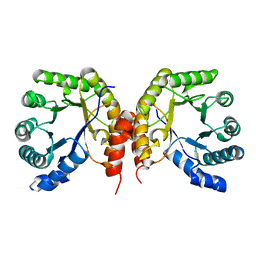 | | Apo structure of a methyltransferase component involved in O-demethylation | | Descriptor: | Dihydropteroate synthase DHPS | | Authors: | Sjuts, H, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2013-12-14 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of the methyltransferase component of Desulfitobacterium hafniense DCB-2 O-demethylase shed light on methyltetrahydrofolate formation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4O1F
 
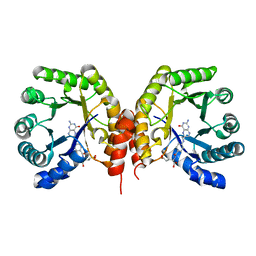 | | Structure of a methyltransferase component in complex with THF involved in O-demethylation | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, Dihydropteroate synthase DHPS | | Authors: | Sjuts, H, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2013-12-15 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the methyltransferase component of Desulfitobacterium hafniense DCB-2 O-demethylase shed light on methyltetrahydrofolate formation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4DQK
 
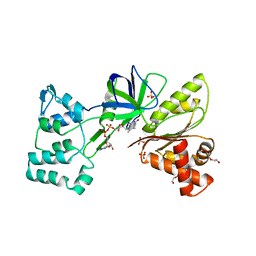 | |
4GU5
 
 | | Structure of Full-length Drosophila Cryptochrome | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Zoltowski, B.D, Vaidya, A.T, Top, D, Widom, J, Young, M.W, Levy, C, Jones, A.R, Scrutton, N.S, Leys, D, Crane, B.R. | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Updated structure of Drosophila cryptochrome.
Nature, 495, 2013
|
|
4EPP
 
 | |
4EP6
 
 | | Crystal structure of the XplA heme domain in complex with imidazole and PEG | | Descriptor: | Cytochrome P450-like protein XplA, IMIDAZOLE, PENTAETHYLENE GLYCOL, ... | | Authors: | Bui, S.H, McLean, K.J, Cheesman, M.R, Bradley, J.M, Rigby, S.E.J, Leys, D, Munro, A.W. | | Deposit date: | 2012-04-17 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unusual Spectroscopic and Ligand Binding Properties of the Cytochrome P450-Flavodoxin Fusion Enzyme XplA.
J.Biol.Chem., 287, 2012
|
|
4O1E
 
 | | Structure of a methyltransferase component in complex with MTHF involved in O-demethylation | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, Dihydropteroate synthase DHPS | | Authors: | Sjuts, H, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2013-12-15 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structures of the methyltransferase component of Desulfitobacterium hafniense DCB-2 O-demethylase shed light on methyltetrahydrofolate formation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4G47
 
 | | Structure of cytochrome P450 CYP121 in complex with 4-(1H-1,2,4-triazol-1-yl)phenol | | Descriptor: | 4-(1H-1,2,4-triazol-1-yl)phenol, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hudson, S.A, McLean, K.J, Surade, S, Yang, Y.-Q, Leys, D, Ciulli, A, Munro, A.W, Abell, C. | | Deposit date: | 2012-07-16 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Application of Fragment Screening and Merging to the Discovery of Inhibitors of the Mycobacterium tuberculosis Cytochrome P450 CYP121
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4G44
 
 | | Structure of P450 CYP121 in complex with lead compound MB286, 3-((1H-1,2,4-triazol-1-yl)methyl)aniline | | Descriptor: | 3-(1H-1,2,4-triazol-1-ylmethyl)aniline, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hudson, S.A, McLean, K.J, Surade, S, Yang, Y.-Q, Leys, D, Ciulli, A, Munro, A.W, Abell, C. | | Deposit date: | 2012-07-16 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Application of Fragment Screening and Merging to the Discovery of Inhibitors of the Mycobacterium tuberculosis Cytochrome P450 CYP121
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4G45
 
 | | Structure of cytochrome CYP121 in complex with 2-methylquinolin-6-amine | | Descriptor: | 2-methylquinolin-6-amine, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hudson, S.A, McLean, K.J, Surade, S, Yang, Y.-Q, Leys, D, Ciulli, A, Munro, A.W, Abell, C. | | Deposit date: | 2012-07-16 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Application of Fragment Screening and Merging to the Discovery of Inhibitors of the Mycobacterium tuberculosis Cytochrome P450 CYP121
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4G46
 
 | | Structure of cytochrome P450 CYP121 in complex with 4-oxo-4,5,6,7-tetrahydrobenzofuran-3-carboxylate | | Descriptor: | 4-oxo-4,5,6,7-tetrahydro-1-benzofuran-3-carboxylic acid, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hudson, S.A, McLean, K.J, Surade, S, Yang, Y.-Q, Leys, D, Ciulli, A, Munro, A.W, Abell, C. | | Deposit date: | 2012-07-16 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Application of Fragment Screening and Merging to the Discovery of Inhibitors of the Mycobacterium tuberculosis Cytochrome P450 CYP121
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3K8D
 
 | | Crystal structure of E. coli lipopolysaccharide specific CMP-KDO synthetase in complex with CTP and 2-deoxy-Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, 3-deoxy-manno-octulosonate cytidylyltransferase, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Heyes, D.J, Levy, C.W, Lafite, P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-10-14 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based mechanism of CMP-2-keto-3-deoxymanno-octulonic acid synthetase: convergent evolution of a sugar-activating enzyme with DNA/RNA polymerases
J.Biol.Chem., 284, 2009
|
|
