3IIE
 
 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis. | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, MAGNESIUM ION | | Authors: | Osipiuk, J, Mulligan, R, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-31 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | X-ray crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis.
To be Published
|
|
3C2Q
 
 | | Crystal structure of conserved putative LOR/SDH protein from Methanococcus maripaludis S2 | | Descriptor: | IMIDAZOLE, NICKEL (II) ION, Uncharacterized conserved protein | | Authors: | Duke, N, Gu, M, Mulligan, R, Conrad, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-25 | | Release date: | 2008-02-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of conserved putative LOR/SDH protein from Methanococcus maripaludis S2
To be Published
|
|
3N70
 
 | | The Crystal Structure of the P-loop NTPase domain of the Sigma-54 transport activator from E. coli to 2.8A | | Descriptor: | SULFATE ION, Transport activator | | Authors: | Stein, A.J, Mulligan, R, Volkart, L, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-26 | | Release date: | 2010-07-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the P-loop NTPase domain of the Sigma-54 transport activator from E. coli to 2.8A
To be Published
|
|
3UHF
 
 | | Crystal Structure of Glutamate Racemase from Campylobacter jejuni subsp. jejuni | | Descriptor: | CHLORIDE ION, D-GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Maltseva, N, Mulligan, R, Kwon, K, Kim, Y, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-03 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Glutamate Racemase
from Campylobacter jejuni subsp. jejuni
To be Published
|
|
3NJL
 
 | | D116A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis, at pH7.5 | | Descriptor: | MAGNESIUM ION, Peptidase | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
3DQG
 
 | | Peptide-binding domain of heat shock 70 kDa protein F, mitochondrial precursor, from Caenorhabditis elegans. | | Descriptor: | Heat shock 70 kDa protein F | | Authors: | Osipiuk, J, Mulligan, R, Gu, M, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | X-ray crystal structure of peptide-binding domain of heat shock 70 kDa protein F, mitochondrial precursor, from Caenorhabditis elegans.
To be Published
|
|
3E0R
 
 | | Crystal structure of cppA protein from Streptococcus pneumoniae TIGR4 | | Descriptor: | C3-degrading proteinase (CppA protein), CHLORIDE ION | | Authors: | Nocek, B, Mulligan, R, Abdullah, J, Otwinowski, Z, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-31 | | Release date: | 2008-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of cppA protein from Streptococcus pneumoniae TIGR4
To be Published
|
|
3NJJ
 
 | | P115A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis | | Descriptor: | Peptidase | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
2R6H
 
 | | Crystal structure of the domain comprising the NAD binding and the FAD binding regions of the NADH:ubiquinone oxidoreductase, Na translocating, F subunit from Porphyromonas gingivalis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH:ubiquinone oxidoreductase, Na translocating, ... | | Authors: | Kim, Y, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-05 | | Release date: | 2007-09-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of the Domain Comprising the Regions Binding NAD and FAD from the NADH:Ubiquinone Oxidoreductase, Na Translocating, F Subunit from Porphyromonas gingivalis.
To be Published
|
|
2NRK
 
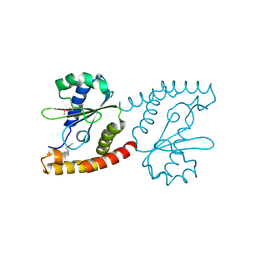 | |
3UHP
 
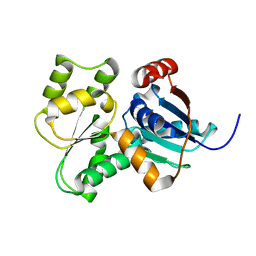 | | Crystal Structure of Glutamate Racemase from Campylobacter jejuni subsp. jejuni | | Descriptor: | Glutamate racemase | | Authors: | Maltseva, N, Mulligan, R, Kwon, K, Kim, Y, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-03 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal Structure of Glutamate Racemase
from Campylobacter jejuni subsp. jejuni
To be Published, 2011
|
|
3BIO
 
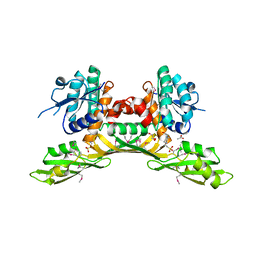 | | Crystal structure of oxidoreductase (Gfo/Idh/MocA family member) from Porphyromonas gingivalis W83 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, GLYCEROL, ... | | Authors: | Nocek, B, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of oxidoreductase (Gfo/Idh/MocA family member) from Porphyromonas gingivalis W83.
To be Published
|
|
3NJI
 
 | | S114A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis. | | Descriptor: | Peptidase | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
3B8B
 
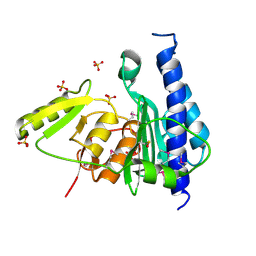 | | Crystal structure of CysQ from Bacteroides thetaiotaomicron, a bacterial member of the inositol monophosphatase family | | Descriptor: | CHLORIDE ION, CysQ, sulfite synthesis pathway protein, ... | | Authors: | Cuff, M.E, Mulligan, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-10-31 | | Release date: | 2007-12-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of CysQ from Bacteroides thetaiotaomicron, a bacterial member of the inositol monophosphatase family.
TO BE PUBLISHED
|
|
3NJF
 
 | | Y26F mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis | | Descriptor: | Peptidase | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
3NJN
 
 | | Q118A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Peptidase | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
2NR5
 
 | | Crystal Structure of Protein of Unknown Function SO2669 from Shewanella oneidensis MR-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Hypothetical protein SO2669 | | Authors: | Nocek, B, Mulligan, R, Peppler, T, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-01 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of SO2669 protein from Shewanella onedensis Mr-1, a singleton of unknown functions.
To be Published
|
|
3E9A
 
 | | Crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, SULFATE ION | | Authors: | Nocek, B, Mulligan, R, Kwon, K, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-08-21 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from Vibrio cholerae O1
biovar eltor str. N16961
To be Published
|
|
3NJM
 
 | | P117A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis. | | Descriptor: | Peptidase | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
3NJG
 
 | | K98A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis. | | Descriptor: | Peptidase | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
3NJK
 
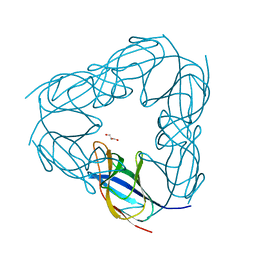 | | D116A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis, at pH5.5 | | Descriptor: | GLYCEROL, Peptidase | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
2O1A
 
 | | Crystal structure of iron-regulated surface determinant protein A from Staphylococcus aureus- targeted domain 47...188 | | Descriptor: | 1,2-ETHANEDIOL, Iron-regulated surface determinant protein A, SULFATE ION | | Authors: | Chang, C, Mulligan, R, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of iron-regulated surface determinant protein A from
Staphylococcus aureus - targeted domain 47...188
To be Published
|
|
3NUA
 
 | | Crystal Structure of Phosphoribosylaminoimidazole-Succinocarboxamide Synthase from Clostridium perfringens | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, CITRIC ACID, ... | | Authors: | Kim, Y, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Phosphoribosylaminoimidazole-Succinocarboxamide Synthase from Clostridium perfringens
To be Published
|
|
3OUU
 
 | | Crystal Structure of Biotin Carboxylase-beta-gamma-ATP Complex from Campylobacter jejuni | | Descriptor: | Biotin carboxylase, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-15 | | Release date: | 2010-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Crystal Structure of Biotin Carboxylase-beta-gamma-ATP Complex from Campylobacter jejuni
TO BE PUBLISHED
|
|
3OUZ
 
 | | Crystal Structure of Biotin Carboxylase-ADP complex from Campylobacter jejuni | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Biotin carboxylase, D-MALATE, ... | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-15 | | Release date: | 2010-10-13 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal Structure of Biotin Carboxylase-ADP complex from Campylobacter jejuni
TO BE PUBLISHED
|
|
