7AQE
 
 | | Structure of SARS-CoV-2 Main Protease bound to UNC-2327 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Meents, A. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6SU4
 
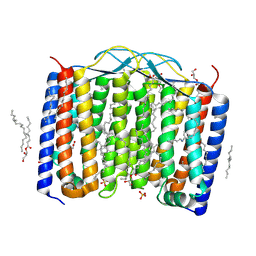 | | Crystal structure of the 48C12 heliorhodopsin in the blue form at pH 4.3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 48C12 heliorhodopsin, ACETATE ION, ... | | Authors: | Kovalev, K, Volkov, D, Astashkin, R, Alekseev, A, Gushchin, I, Gordeliy, V. | | Deposit date: | 2019-09-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structural insights into the heliorhodopsin family.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6SU3
 
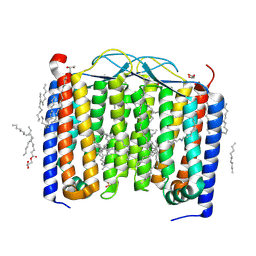 | | Crystal structure of the 48C12 heliorhodopsin in the violet form at pH 8.8 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 48C12 heliorhodopsin, EICOSANE, ... | | Authors: | Kovalev, K, Volkov, D, Astashkin, R, Alekseev, A, Gushchin, I, Gordeliy, V. | | Deposit date: | 2019-09-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structural insights into the heliorhodopsin family.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7ZOU
 
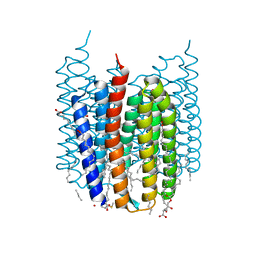 | | Crystal structure of Synechocystis halorhodopsin (SyHR), Cl-pumping mode, ground state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, EICOSANE, ... | | Authors: | Kovalev, K, Bukhdruker, S, Astashkin, R, Vaganova, S, Gordeliy, V. | | Deposit date: | 2022-04-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insights into light-driven anion pumping in cyanobacteria.
Nat Commun, 13, 2022
|
|
7ZOW
 
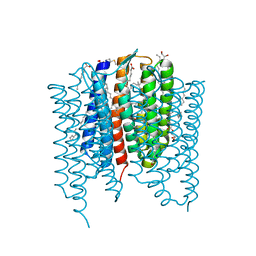 | | Crystal structure of Synechocystis halorhodopsin (SyHR), Cl-pumping mode, O state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Bukhdruker, S, Astashkin, R, Vaganova, S, Gordeliy, V. | | Deposit date: | 2022-04-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into light-driven anion pumping in cyanobacteria.
Nat Commun, 13, 2022
|
|
7ZOV
 
 | | Crystal structure of Synechocystis halorhodopsin (SyHR), Cl-pumping mode, K state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, EICOSANE, ... | | Authors: | Kovalev, K, Bukhdruker, S, Astashkin, R, Vaganova, S, Gordeliy, V. | | Deposit date: | 2022-04-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into light-driven anion pumping in cyanobacteria.
Nat Commun, 13, 2022
|
|
7ZOY
 
 | | Crystal structure of Synechocystis halorhodopsin (SyHR), SO4-bound form, ground state | | Descriptor: | CHLORIDE ION, EICOSANE, GLYCEROL, ... | | Authors: | Kovalev, K, Bukhdruker, S, Astashkin, R, Vaganova, S, Gordeliy, V. | | Deposit date: | 2022-04-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural insights into light-driven anion pumping in cyanobacteria.
Nat Commun, 13, 2022
|
|
4U4M
 
 | | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate | | Descriptor: | 1,2-ETHANEDIOL, PYRUVIC ACID, UREA, ... | | Authors: | Manoj Kumar, P, Bhaskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate
To be published
|
|
1OGY
 
 | | Crystal structure of the heterodimeric nitrate reductase from Rhodobacter sphaeroides | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DIHEME CYTOCHROME C NAPB MOLECULE: NITRATE REDUCTASE, HEME C, ... | | Authors: | Arnoux, P, Sabaty, M, Alric, J, Frangioni, B, Guigliarelli, B, Adriano, J.-M, Pignol, D. | | Deposit date: | 2003-05-19 | | Release date: | 2003-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Redox Plasticity in the Heterodimeric Periplasmic Nitrate Reductase
Nat.Struct.Biol., 10, 2003
|
|
1ELR
 
 | | Crystal structure of the TPR2A domain of HOP in complex with the HSP90 peptide MEEVD | | Descriptor: | HSP90-PEPTIDE MEEVD, NICKEL (II) ION, TPR2A-DOMAIN OF HOP | | Authors: | Scheufler, C, Brinker, A, Hartl, F.U, Moarefi, I. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-26 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine.
Cell(Cambridge,Mass.), 101, 2000
|
|
1ELW
 
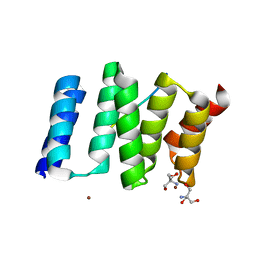 | | Crystal structure of the TPR1 domain of HOP in complex with a HSC70 peptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HSC70-PEPTIDE, NICKEL (II) ION, ... | | Authors: | Scheufler, C, Brinker, A, Hartl, F.U, Moarefi, I. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine
Cell(Cambridge,Mass.), 101, 2000
|
|
4OE7
 
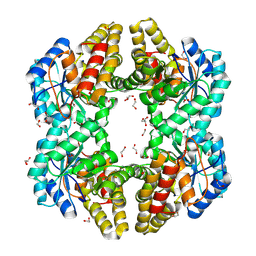 | | Crystal structure of YagE, a KDG aldolase protein, in complex with aldol condensed product of pyruvate and glyoxal | | Descriptor: | (4R)-4-hydroxy-2,5-dioxopentanoic acid, (4S)-4-hydroxy-2,5-dioxopentanoic acid, 1,2-ETHANEDIOL, ... | | Authors: | Manoj Kumar, P, Baskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-01-12 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of YagE, a KDG aldolase protein, in complex with aldol condensed product of pyruvate and glyoxal
To be Published
|
|
4ONV
 
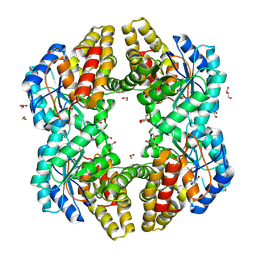 | | Crystal structure of YagE, a KDG aldolase protein in complex with 2-Keto-3-deoxy gluconate | | Descriptor: | 1,2-ETHANEDIOL, 2-KETO-3-DEOXYGLUCONATE, GLYCEROL, ... | | Authors: | Manoj Kumar, P, Bhaskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-01-29 | | Release date: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structure of YagE, a KDG aldolase protein in complex with 2-Keto-3-deoxy gluconate
To be Published
|
|
7A42
 
 | |
7A43
 
 | |
7A44
 
 | | CO-bound sperm whale myoglobin measured by serial synchrotron crystallography | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mehrabi, P, Schulz, E.C, Buecker, R. | | Deposit date: | 2020-08-19 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Serial femtosecond and serial synchrotron crystallography can yield data of equivalent quality: A systematic comparison.
Sci Adv, 7, 2021
|
|
7A45
 
 | | CO-bound sperm whale myoglobin measured by serial femtosecond crystallography | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mehrabi, P, Schulz, E.C, Buecker, R. | | Deposit date: | 2020-08-19 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Serial femtosecond and serial synchrotron crystallography can yield data of equivalent quality: A systematic comparison.
Sci Adv, 7, 2021
|
|
6RNC
 
 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Lysozyme with GlcNAc3 - 100ms diffusion time. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | Deposit date: | 2019-05-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
6RNB
 
 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Lysozyme with GlcNAc3 50ms diffusion time | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | Deposit date: | 2019-05-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
6RNF
 
 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Xylose Isomerase 30 ms timepoint | | Descriptor: | MAGNESIUM ION, Xylose isomerase, alpha-D-glucopyranose | | Authors: | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | Deposit date: | 2019-05-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
6RND
 
 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Xylose Isomerase 15 ms timepoint | | Descriptor: | MAGNESIUM ION, Xylose isomerase, alpha-D-glucopyranose | | Authors: | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | Deposit date: | 2019-05-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
6ZWZ
 
 | |
6ZWY
 
 | |
6ZX3
 
 | | OMPD-domain of human UMPS in complex with 6-thiocarboxamido-UMP at 1.15 Angstroms resolution | | Descriptor: | GLYCEROL, PROLINE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Schimdt, T. | | Deposit date: | 2020-07-29 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6ZX1
 
 | | OMPD-domain of human UMPS in complex with 6-Aza-UMP at 1.0 Angstroms resolution | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, PROLINE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-07-29 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
