8XWX
 
 | |
9ARD
 
 | |
7LB7
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A.Y, Kneller, D.W, Coates, L. | | Deposit date: | 2021-01-07 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Direct Observation of Protonation State Modulation in SARS-CoV-2 Main Protease upon Inhibitor Binding with Neutron Crystallography.
J.Med.Chem., 64, 2021
|
|
8B0W
 
 | |
4FE1
 
 | | Improving the Accuracy of Macromolecular Structure Refinement at 7 A Resolution | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Fromme, R, Adams, P.D, Fromme, P, Levitt, M, Schroeder, G.F, Brunger, A.T. | | Deposit date: | 2012-05-29 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.9228 Å) | | Cite: | Improving the accuracy of macromolecular structure refinement at 7 A resolution.
Structure, 20, 2012
|
|
7JIZ
 
 | |
7JMC
 
 | | Sheep Connexin-50 at 2.5 angstroms resolution, Lipid Class 3 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7KDF
 
 | | Structure of Stu2 Bound to dwarf Ndc80c | | Descriptor: | NDC80 isoform 1,NDC80 isoform 1, NUF2 isoform 1,NUF2 isoform 1, SPC25 isoform 1,SPC25 isoform 1, ... | | Authors: | Zahm, J.A, Stewart, M.G, Miller, M.P, Harrison, S.C. | | Deposit date: | 2020-10-08 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis of Stu2 recruitment to yeast kinetochores.
Elife, 10, 2021
|
|
5WEY
 
 | | Joint X-ray/neutron structure of Concanavalin A with alpha1-2 D-mannobiose | | Descriptor: | CALCIUM ION, Concanavalin-A, MANGANESE (II) ION, ... | | Authors: | Kovalevsky, A, Gerlits, O.O, Woods, R.J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | Mannobiose Binding Induces Changes in Hydrogen Bonding and Protonation States of Acidic Residues in Concanavalin A As Revealed by Neutron Crystallography.
Biochemistry, 56, 2017
|
|
7SBG
 
 | |
7SD2
 
 | |
5H33
 
 | |
8C3W
 
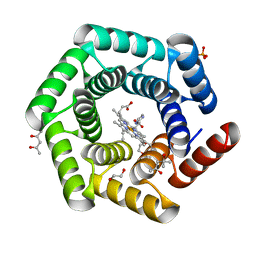 | | Crystal structure of a computationally designed heme binding protein, dnHEM1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ortmayer, M, Levy, C. | | Deposit date: | 2022-12-29 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Heme Enzymes with a Tunable Substrate Binding Pocket Adjacent to an Open Metal Coordination Site.
J.Am.Chem.Soc., 145, 2023
|
|
8QI7
 
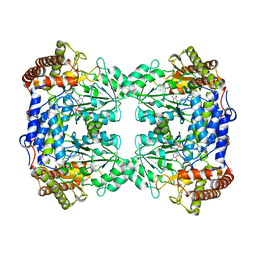 | |
8QY4
 
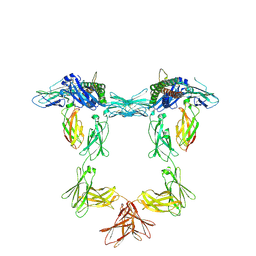 | | Structure of interleukin 11 (gp130 P496L mutant). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-11, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural insights into IL-11-mediated signalling and human IL6ST variant-associated immunodeficiency.
Nat Commun, 15, 2024
|
|
8QY5
 
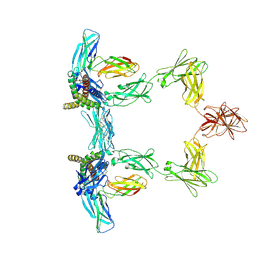 | | Structure of interleukin 6. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-6, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into IL-11-mediated signalling and human IL6ST variant-associated immunodeficiency.
Nat Commun, 15, 2024
|
|
8QY6
 
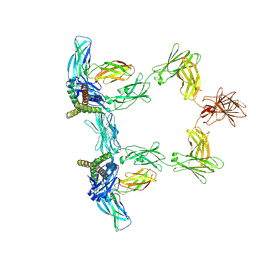 | | Structure of interleukin 6 (gp130 P496L mutant). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-6, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural insights into IL-11-mediated signalling and human IL6ST variant-associated immunodeficiency.
Nat Commun, 15, 2024
|
|
8Q6B
 
 | | The RSL-D32N - sulfonato-calix[8]arene complex, I23 form, citrate pH 4.0, obtained by cross-seeding | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Flood, R.J, Crowley, P.B. | | Deposit date: | 2023-08-11 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Supramolecular Synthons in Protein-Ligand Frameworks.
Cryst.Growth Des., 24, 2024
|
|
8QSQ
 
 | |
8QBF
 
 | | Compact state - Pil1 dimer with lipid headgroups fitted in native eisosome lattice bound to plasma membrane microdomain | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PHOSPHOSERINE, Sphingolipid long chain base-responsive protein PIL1 | | Authors: | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 632, 2024
|
|
8QB7
 
 | |
8QB8
 
 | |
6RTE
 
 | | Dihydro-heme d1 dehydrogenase NirN in complex with DHE | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cytochrome c, HEME C | | Authors: | Kluenemann, T, Preuss, A, Layer, G, Blankenfeldt, W. | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Dihydro-Heme d1Dehydrogenase NirN from Pseudomonas aeruginosa Reveals Amino Acid Residues Essential for Catalysis.
J.Mol.Biol., 431, 2019
|
|
8QBD
 
 | | Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol, PI(4,5)P2 35:20:20:15:10) | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, Sphingolipid long chain base-responsive protein PIL1 | | Authors: | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 632, 2024
|
|
8QBG
 
 | |
