5QTU
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(3R,7S)-7-{[1-(3-chloro-2-fluorophenyl)-5-methyl-1H-imidazole-4-carbonyl]amino}-3-methyl-2-oxo-2,3,4,5,6,7-hexahydro-1H-12,8-(metheno)-1,9-benzodiazacyclotetradecin-15-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2019-10-16 | | Release date: | 2019-12-25 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Potent, Orally Bioavailable and Efficacious Macrocyclic Inhibitors of Factor XIa. Discovery of Pyridine-Based Macrocycles Possessing Phenylazole Carboxamide P1 Groups.
J.Med.Chem., 63, 2019
|
|
5QTW
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl (2R,7S)-7-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-14-[(methoxycarbonyl)amino]-2,3,4,5,6,7-hexahydro-1H-8,11-epimino-1,9-benzodiazacyclotridecine-2-carboxylate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-29 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Orally bioavailable amine-linked macrocyclic inhibitors of factor XIa.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5QTV
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(2R,7S)-7-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-2-(trifluoromethyl)-2,3,4,5,6,7-hexahydro-1H-8,11-epimino-1,9-benzodiazacyclotridecin-14-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-29 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Orally bioavailable amine-linked macrocyclic inhibitors of factor XIa.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5QTX
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR ethyl (2R,7S)-7-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-14-[(methoxycarbonyl)amino]-1,2,3,4,5,6,7,9-octahydro-11,8-(azeno)-1,9-benzodiazacyclotridecine-2-carboxylate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-29 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Orally bioavailable amine-linked macrocyclic inhibitors of factor XIa.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5QTY
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR ethyl (2R,7S)-7-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-15-[(methoxycarbonyl)amino]-2,3,4,5,6,7-hexahydro-1H-12,8-(metheno)-1,9-benzodiazacyclotetradecine-2-carboxylate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Orally bioavailable amine-linked macrocyclic inhibitors of factor XIa.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
4KOM
 
 | | The structure of hemagglutinin from avian-origin H7N9 influenza virus in complex with avian receptor analog 3'SLNLN (NeuAcα2-3Galβ1-4GlcNAcβ1-3Galβ1-4Glc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2, ... | | Authors: | Shi, Y, Zhang, W, Wang, F, Qi, J, Song, H, Wu, Y, Gao, F, Zhang, Y, Fan, Z, Gong, W, Wang, D, Shu, Y, Wang, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-05-12 | | Release date: | 2013-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses.
Science, 342, 2013
|
|
4KON
 
 | | The structure of hemagglutinin from avian-origin H7N9 influenza virus in complex with human receptor analog 6'SLNLN (NeuAcα2-6Galβ1-4GlcNAcβ1-3Galβ1-4Glc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2, ... | | Authors: | Shi, Y, Zhang, W, Wang, F, Qi, J, Song, H, Wu, Y, Gao, F, Zhang, Y, Fan, Z, Gong, W, Wang, D, Shu, Y, Wang, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-05-12 | | Release date: | 2013-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses.
Science, 342, 2013
|
|
4KOL
 
 | | The structure of hemagglutinin from avian-origin H7N9 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2 | | Authors: | Shi, Y, Zhang, W, Wang, F, Qi, J, Song, H, Wu, Y, Gao, F, Zhang, Y, Fan, Z, Gong, W, Wang, D, Shu, Y, Wang, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-05-12 | | Release date: | 2013-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses.
Science, 342, 2013
|
|
5QCK
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR 4-[[(2~{S},3~{R})-1-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3-phenyl-pyrrolidin-2-yl]carbonylamino]benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-[[(2~{S},3~{R})-1-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3-phenyl-pyrrolidin-2-yl]carbonylamino]benzoic acid, Coagulation factor XI, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-08 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212).
J. Med. Chem., 60, 2017
|
|
5QCM
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl ~{N}-[4-[[(1~{S})-2-[(~{E})-3-[3-chloranyl-2-fluoranyl-6-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]phenyl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-08 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212).
J. Med. Chem., 60, 2017
|
|
5QCL
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid, Coagulation factor XI, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-08 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212).
J. Med. Chem., 60, 2017
|
|
5QCN
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-5-[(3~{S})-3-ethoxycarbonylpiperidin-1-yl]carbonyl-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-5-[(3~{S})-3-ethoxycarbonylpiperidin-1-yl]carbonyl-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid, Coagulation factor XI, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-08 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212).
J. Med. Chem., 60, 2017
|
|
5H65
 
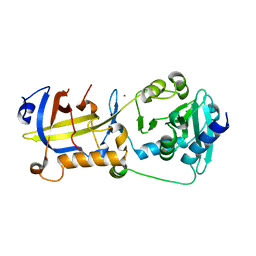 | | Crystal structure of human POT1 and TPP1 | | Descriptor: | Adrenocortical dysplasia protein homolog, Protection of telomeres protein 1, ZINC ION | | Authors: | Chen, C, Wu, J, Lei, M. | | Deposit date: | 2016-11-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into POT1-TPP1 interaction and POT1 C-terminal mutations in human cancer.
Nat Commun, 8, 2017
|
|
5QQP
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(5E,8S)-8-[(4S)-4-(3-chlorophenyl)-2-oxopiperidin-1-yl]-2-oxo-1,3,4,7,8,10-hexahydro-2H-12,9-(azeno)-1,10-benzodiazacyclotetradecin-15-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-18 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure based design of macrocyclic factor XIa inhibitors: Discovery of cyclic P1 linker moieties with improved oral bioavailability.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
5QQO
 
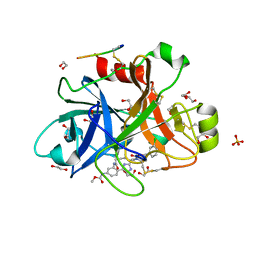 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(5E,8S)-8-[(6R)-6-(3-chlorophenyl)-2-oxo-1,3-oxazinan-3-yl]-2-oxo-1,3,4,7,8,10-hexahydro-2H-12,9-(azeno)-1,10-benzodiazacyclotetradecin-15-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, MET-ASP-ASP-ASP-ASP-LYS-MET-ASP-ASN-GLU-CYS-THR-THR-LYS-ILE-LYS-PRO-ARG, ... | | Authors: | Sheriff, S. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-18 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure based design of macrocyclic factor XIa inhibitors: Discovery of cyclic P1 linker moieties with improved oral bioavailability.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
3HH8
 
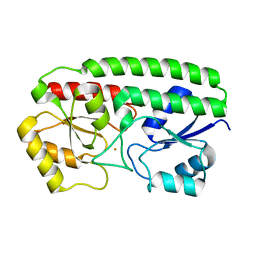 | | Crystal Structure and metal binding properties of the lipoprotein MtsA | | Descriptor: | FE (III) ION, Metal ABC transporter substrate-binding lipoprotein | | Authors: | Baker, E.N, Baker, H.M, Sun, X, Ye, Q.-Y. | | Deposit date: | 2009-05-15 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure and metal binding properties of the lipoprotein MtsA, responsible for iron transport in Streptococcus pyogenes.
Biochemistry, 48, 2009
|
|
5Q0G
 
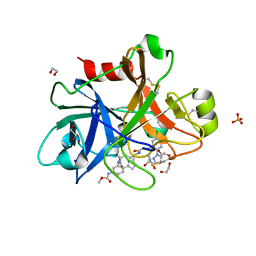 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(3R,7S)-7-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-3-ethyl-2-oxo-1,2,3,4,5,6,7,9-octahydro-11,8-(azeno)-1,9-benzodiazacyclotridecin-14-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Macrocyclic inhibitors of Factor XIa: Discovery of alkyl-substituted macrocyclic amide linkers with improved potency.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5Q0D
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(7S)-7-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-2-oxo-1,2,3,4,5,6,7,9-octahydro-11,8-(azeno)-1,9-benzodiazacyclotridecin-14-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-12 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Macrocyclic inhibitors of Factor XIa: Discovery of alkyl-substituted macrocyclic amide linkers with improved potency.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5Q0H
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(4R,5E,8S)-11-chloro-8-[(2,6-difluoro-4-methylbenzene-1-carbonyl)amino]-4-methyl-2-oxo-1,3,4,7,8,10-hexahydro-2H-12,9-(azeno)-1,10-benzodiazacyclotetradecin-15-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-12 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Macrocyclic inhibitors of Factor XIa: Discovery of alkyl-substituted macrocyclic amide linkers with improved potency.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5Q0F
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(4R,5E,8S)-8-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-4-methyl-2-oxo-1,3,4,7,8,10-hexahydro-2H-12,9-(azeno)-1,10-benzodiazacyclotetradecin-15-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, MET-ASP-ASP-ASP-ASP-LYS-MET-ASP-ASN-GLU-CYS-THR-THR-LYS-ILE-LYS-PRO-ARG, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Macrocyclic inhibitors of Factor XIa: Discovery of alkyl-substituted macrocyclic amide linkers with improved potency.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5Q0E
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(4S,8S)-8-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-4-methyl-2-oxo-1,3,4,5,6,7,8,10-octahydro-2H-12,9-(azeno)-1,10-benzodiazacyclotetradecin-15-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-12 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Macrocyclic inhibitors of Factor XIa: Discovery of alkyl-substituted macrocyclic amide linkers with improved potency.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6C0S
 
 | |
2XEM
 
 | | Induced-fit and allosteric effects upon polyene binding revealed by crystal structures of the Dynemicin thioesterase | | Descriptor: | (3E,5E,7E,9E,11E,13E)-pentadeca-3,5,7,9,11,13-hexaen-2-one, DYNE7 | | Authors: | Liew, C.W, Sharff, A, Kotaka, M, Kong, R, Bricogne, G, Liang, Z.X, Lescar, J. | | Deposit date: | 2010-05-17 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induced-Fit Upon Ligand Binding Revealed by Crystal Structures of the Hot-Dog Fold Thioesterase in Dynemicin Biosynthesis.
J.Mol.Biol., 404, 2010
|
|
8Y8E
 
 | | Structure of HCoV-HKU1C spike in the inactive-2up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y87
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-1up conformation with 1TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
