5LBV
 
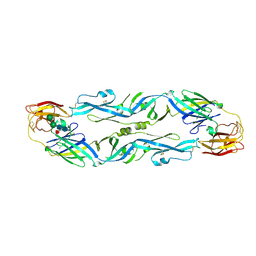 | | Structural basis of zika and dengue virus potent antibody cross-neutralization | | Descriptor: | SODIUM ION, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, envelope protein E | | Authors: | Barba-Spaeth, G. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of potent Zika-dengue virus antibody cross-neutralization.
Nature, 536, 2016
|
|
5LBS
 
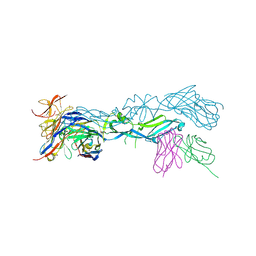 | | structural basis of Zika and Dengue virus potent antibody cross-neutralization | | Descriptor: | 1,2-ETHANEDIOL, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C8, SULFATE ION, ... | | Authors: | Vaney, M.C, Rouvinski, A, Barba-Spaeth, G, Rey, F.A. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis of potent Zika-dengue virus antibody cross-neutralization.
Nature, 536, 2016
|
|
2KXM
 
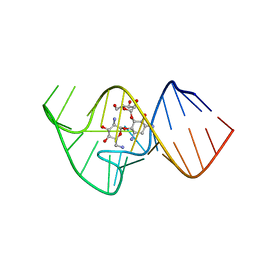 | | Solution NMR Structure of the 27 nucleotide engineered neomycin sensing riboswitch RNA-ribostmycin complex | | Descriptor: | RIBOSTAMYCIN, RNA (27-MER) | | Authors: | Duchardt-Ferner, E, Weigand, J.E, Ohlenschlager, O, Schmidtke, S.R, Suess, B, Wohnert, J. | | Deposit date: | 2010-05-10 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Highly modular structure and ligand binding by conformational capture in a minimalistic riboswitch.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
6PH4
 
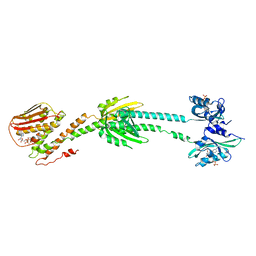 | | Full length LOV-PAS-HK construct from the LOV-HK sensory protein from Brucella abortus (light-adapted, construct 15-489) | | Descriptor: | Blue-light-activated histidine kinase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
1G0V
 
 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH A IA3 MUTANT, MVV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEASE A INHIBITOR 3, PROTEINASE A, ... | | Authors: | Phylip, L.H, Lees, W, Brownsey, B.G, Bur, D, Dunn, B.M, Winther, J, Gustchina, A, Li, M, Copeland, T, Wlodawer, A, Kay, J. | | Deposit date: | 2000-10-09 | | Release date: | 2001-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The potency and specificity of the interaction between the IA3 inhibitor and its target aspartic proteinase from Saccharomyces cerevisiae.
J.Biol.Chem., 276, 2001
|
|
1GFW
 
 | |
3T6I
 
 | | Endothiapepsin in complex with an azepin derivative | | Descriptor: | (3R)-3-({(4-aminobenzyl)[(4-aminophenyl)acetyl]amino}methyl)-5-(hydroxymethyl)-2,3,4,7-tetrahydro-1H-azepinium, (3R)-3-({(4-aminobenzyl)[(4-aminophenyl)acetyl]amino}methyl)-5-{[(4-bromobenzoyl)oxy]methyl}-2,3,4,7-tetrahydro-1H-azepinium, 4-bromobenzoic acid, ... | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Endothiapepsin in complex with an azepin derivative
To be Published
|
|
1O63
 
 | |
1O6C
 
 | |
1LX6
 
 | |
3VAC
 
 | |
1O6B
 
 | |
1O66
 
 | |
1O67
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yiiM | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O60
 
 | | Crystal structure of KDO-8-phosphate synthase | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O6D
 
 | | Crystal structure of a hypothetical protein | | Descriptor: | Hypothetical UPF0247 protein TM0844 | | Authors: | Structural GenomiX | | Deposit date: | 2003-11-03 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O62
 
 | |
1O65
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yiiM | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O61
 
 | | Crystal structure of a PLP-dependent enzyme with PLP | | Descriptor: | ACETATE ION, PYRIDOXAL-5'-PHOSPHATE, aminotransferase | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O68
 
 | |
1O69
 
 | | Crystal structure of a PLP-dependent enzyme | | Descriptor: | (2-AMINO-4-FORMYL-5-HYDROXY-6-METHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, BETA-MERCAPTOETHANOL, aminotransferase | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O64
 
 | |
3W9E
 
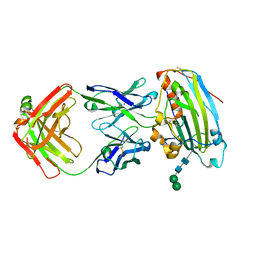 | | Structure of Human Monoclonal Antibody E317 Fab Complex with HSV-2 gD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab heavy chain, Antibody Fab light chain, ... | | Authors: | Lee, C.C, Lin, L.L, Wang, A.H.J. | | Deposit date: | 2013-04-03 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the antibody neutralization of herpes simplex virus
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3W9D
 
 | |
1OQD
 
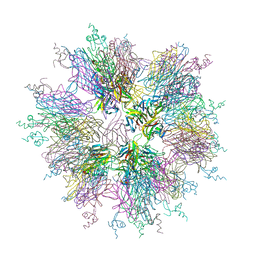 | | Crystal structure of sTALL-1 and BCMA | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, soluble form, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Zhang, G. | | Deposit date: | 2003-03-07 | | Release date: | 2003-05-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ligand-receptor binding revealed by the TNF family member TALL-1.
Nature, 423, 2003
|
|
