8ZRN
 
 | | Structure of abt | | Descriptor: | 5-[[(2~{S})-azetidin-2-yl]methoxy]-2-chloranyl-pyridine, Neuronal acetylcholine receptor subunit alpha-6, Neuronal acetylcholine receptor subunit beta-4,Soluble cytochrome b562, ... | | Authors: | Su, J, Yu, Z, Zhao, Y. | | Deposit date: | 2024-06-04 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Molecular insights into the alpha 6 beta 4 nicotinic acetylcholine receptor function and ligand recognition.
Nat Commun, 16, 2025
|
|
8ZRP
 
 | | Structure of nico | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, Neuronal acetylcholine receptor subunit alpha-6, Neuronal acetylcholine receptor subunit beta-4,Soluble cytochrome b562, ... | | Authors: | Su, J, Yu, Z, Zhao, Y. | | Deposit date: | 2024-06-05 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Molecular insights into the alpha 6 beta 4 nicotinic acetylcholine receptor function and ligand recognition.
Nat Commun, 16, 2025
|
|
9M3O
 
 | |
9M3P
 
 | |
3CO9
 
 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-{3-[4-hydroxy-1-(3-methylbutyl)-2-oxo-1,2-dihydropyrrolo[1,2-b]pyridazin-3-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7 -yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | Deposit date: | 2008-03-27 | | Release date: | 2009-02-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyrrolo[1,2-b]pyridazin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
8RC0
 
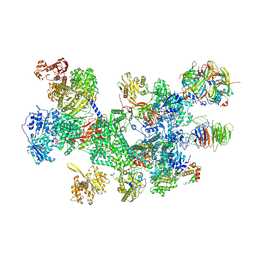 | | Structure of the human 20S U5 snRNP | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Schneider, S, Galej, W.P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human 20S U5 snRNP.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HLC
 
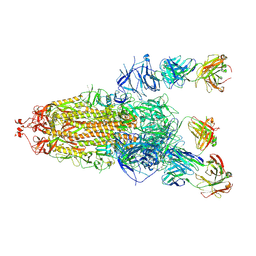 | | S protein of SARS-CoV-2 in complex with 3711 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Y.Y, Guo, Y.Y, Zhou, Q. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-05 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Defining the features and structure of neutralizing antibody targeting the silent face of the SARS-CoV-2 spike N-terminal domain.
MedComm (2020), 5, 2024
|
|
8HLD
 
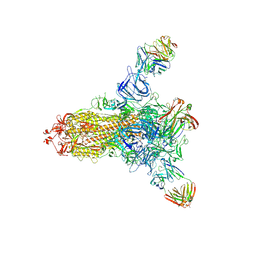 | | S protein of SARS-CoV-2 in complex with 26434 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhang, Y.Y, Guo, Y.Y, Zhou, Q. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-05 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Defining the features and structure of neutralizing antibody targeting the silent face of the SARS-CoV-2 spike N-terminal domain.
MedComm (2020), 5, 2024
|
|
7F5W
 
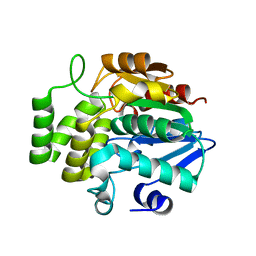 | |
9M3R
 
 | |
9M3T
 
 | |
9M3U
 
 | | Crystal structure of human pyruvate dehydrogenase kinase isoform 2 in complex with ATP competitive inhibitor 24 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyano-N-[(3R)-1-[(3S)-3-ethyl-7-(ethylamino)-5-fluoranyl-2-oxidanylidene-1H-indol-3-yl]piperidin-3-yl]-2-azaspiro[3.3]heptane-6-carboxamide, GLYCEROL, ... | | Authors: | Xu, Z.H, Chen, S, Han, L. | | Deposit date: | 2025-03-03 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of ATP competitive PDHK1/2 dual inhibitors.
Bioorg.Med.Chem.Lett., 122, 2025
|
|
7JUO
 
 | | CBP bromodomain complexed with YF2-23 | | Descriptor: | CREB-binding protein, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-(1-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-3-yl)-1H-indol-4-yl}ethanesulfonamide | | Authors: | Ratia, K.M, Xiong, R, Principe, D, Li, Y, Huang, F, Rana, A, Thatcher, G. | | Deposit date: | 2020-08-20 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | XP-524 is a dual-BET/EP300 inhibitor that represses oncogenic KRAS and potentiates immune checkpoint inhibition in pancreatic cancer.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5GSV
 
 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 142-5 | | Descriptor: | 10-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GR7
 
 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 37-1 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-D alpha chain, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-08 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GSR
 
 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide I5A | | Descriptor: | 9-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GSB
 
 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 37-3 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-D alpha chain, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
6LPF
 
 | | The crystal structure of human cytoplasmic LRS | | Descriptor: | 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, 5'-O-(L-leucylsulfamoyl)adenosine, GLYCEROL, ... | | Authors: | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | Deposit date: | 2020-01-10 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
3CDE
 
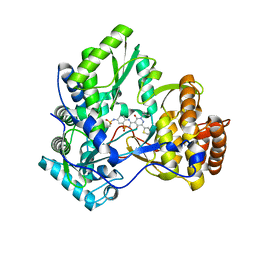 | | Crystal structure of HCV NS5B polymerase with a novel Pyridazinone inhibitor | | Descriptor: | N-{3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-thiophen-2-yl-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2008-02-26 | | Release date: | 2009-03-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel HCV NS5B polymerase inhibitors derived from 4-(1',1'-dioxo-1',4'-dihydro-1'lambda(6)-benzo[1',2',4']thiadiazin-3'-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 3: Further optimization of the 2-, 6-, and 7'-substituents and initial pharmacokinetic assessments.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5WSH
 
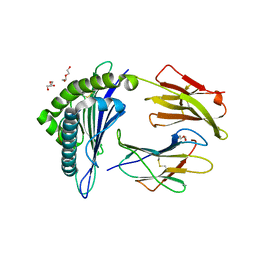 | | Structure of HLA-A2 P130 | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, GLY-VAL-TRP-ILE-ARG-THR-PRO-THR-ALA, ... | | Authors: | Zhang, Y, Wu, Y, Qi, J, Liu, J, Gao, G.F, Meng, S. | | Deposit date: | 2016-12-07 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CD8+T-Cell Response-Associated Evolution of Hepatitis B Virus Core Protein and Disease Progress.
J. Virol., 92, 2018
|
|
3GYN
 
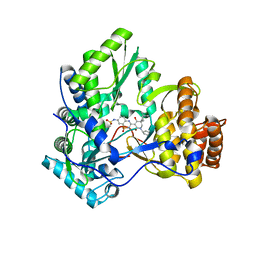 | | Crystal structure of HCV NS5B polymerase with a novel monocyclic dihydropyridinone inhibitor | | Descriptor: | N-{3-[(5R)-1-cyclopentyl-4-hydroxy-5-methyl-5-(3-methylbutyl)-2-oxo-1,2,5,6-tetrahydropyridin-3-yl]-1,1-dioxido-4H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2009-04-04 | | Release date: | 2009-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 5,5'- and 6,6'-dialkyl-5,6-dihydro-1H-pyridin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3IGV
 
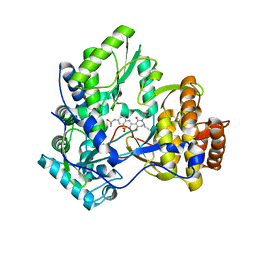 | | Crystal structure of HCV NS5B polymerase with a novel monocyclic dihydro-pyridinone inhibitor | | Descriptor: | N-{3-[(6S)-6-ethyl-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-1,2,5,6-tetrahydropyridin-3-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-DIRECTED RNA POLYMERASE | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2009-07-28 | | Release date: | 2009-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 5,5'- and 6,6'-dialkyl-5,6-dihydro-1H-pyridin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6K13
 
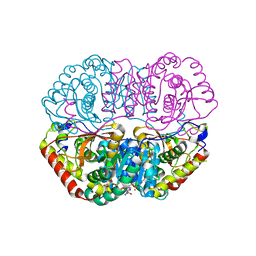 | | Crystal Structure Basis for BmLDH Complex | | Descriptor: | L-lactate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID | | Authors: | Long, Y, Shen, Z. | | Deposit date: | 2019-05-09 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures ofBabesia microtilactate dehydrogenase BmLDH reveal a critical role for Arg99 in catalysis.
Faseb J., 33, 2019
|
|
6J9D
 
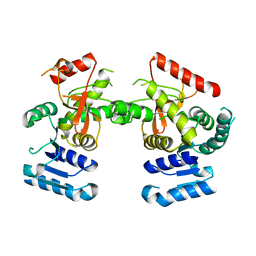 | | Babesia microti lactate dehydrogenase R99A (BmLDHR99A) | | Descriptor: | L-lactate dehydrogenase | | Authors: | Yu, L. | | Deposit date: | 2019-01-22 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Crystal structures ofBabesia microtilactate dehydrogenase BmLDH reveal a critical role for Arg99 in catalysis.
Faseb J., 33, 2019
|
|
6K12
 
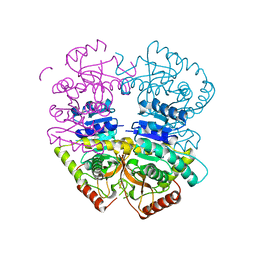 | | Babesia microti lactate dehydrogenase apo form (BmLDH) | | Descriptor: | L-lactate dehydrogenase | | Authors: | Long, Y. | | Deposit date: | 2019-05-09 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal structures ofBabesia microtilactate dehydrogenase BmLDH reveal a critical role for Arg99 in catalysis.
Faseb J., 33, 2019
|
|
