3D6J
 
 | | Crystal structure of Putative haloacid dehalogenase-like hydrolase from Bacteroides fragilis | | Descriptor: | GLYCEROL, PHOSPHATE ION, Putative haloacid dehalogenase-like hydrolase | | Authors: | Chang, C, Wu, R, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-19 | | Release date: | 2008-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Putative haloacid dehalogenase-like hydrolase from Bacteroides fragilis
To be Published
|
|
3CEX
 
 | | Crystal structure of the conserved protein of locus EF_3021 from Enterococcus faecalis | | Descriptor: | ACETIC ACID, GLYCEROL, Uncharacterized protein | | Authors: | Cuff, M.E, Wu, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-29 | | Release date: | 2008-05-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the conserved protein of locus EF_3021 from Enterococcus faecalis.
TO BE PUBLISHED
|
|
5I47
 
 | | Crystal structure of RimK domain protein ATP-grasp from Sphaerobacter thermophilus DSM 20745 | | Descriptor: | GLYCEROL, RimK domain protein ATP-grasp | | Authors: | Chang, C, Duke, N, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-02-11 | | Release date: | 2016-03-16 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of RimK domain protein ATP-grasp from Sphaerobacter thermophilus DSM 20745
To Be Published
|
|
3JUW
 
 | |
4ZTK
 
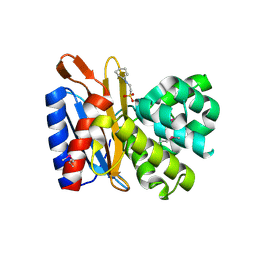 | | Transpeptidase domain of FtsI4 D,D-transpeptidase from Legionella pneumophila. | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Cell division protein FtsI/penicillin binding protein 2 | | Authors: | CUFF, M, OSIPIUK, J, WU, R, ENDRES, M, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-14 | | Release date: | 2015-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Transpeptidase domain of FtsI4 D,D-transpeptidase from Legionella pneumophila.
to be published
|
|
3B85
 
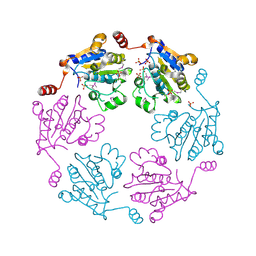 | |
3BED
 
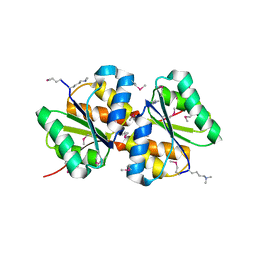 | |
4YCS
 
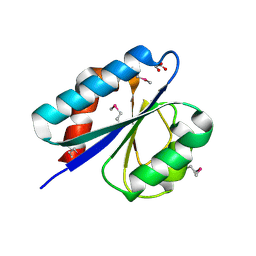 | | Crystal structure of putative lipoprotein from Peptoclostridium difficile 630 (fragment) | | Descriptor: | ACETATE ION, GLYCEROL, SODIUM ION, ... | | Authors: | Michalska, K, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-20 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of putative lipoprotein from Peptoclostridium difficile 630 (fragment)
To Be Published
|
|
3MXQ
 
 | |
3MUQ
 
 | |
5TTX
 
 | |
3OOO
 
 | | The structure of a proline dipeptidase from Streptococcus agalactiae 2603V | | Descriptor: | Proline dipeptidase | | Authors: | Fan, Y, Wu, R, Morales, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The structure of a proline dipeptidase from Streptococcus agalactiae 2603V
To be Published
|
|
3GDZ
 
 | | Crystal structure of arginyl-tRNA synthetase from Klebsiella pneumoniae subsp. pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, Arginyl-tRNA synthetase | | Authors: | Chang, C, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of arginyl-tRNA synthetase from Klebsiella pneumoniae subsp. pneumoniae
To be Published
|
|
3H9P
 
 | | Crystal structure of putative triphosphoribosyl-dephospho-coA synthase from Archaeoglobus fulgidus | | Descriptor: | CHLORIDE ION, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Chang, C, Wu, R, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-30 | | Release date: | 2009-05-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of putative triphosphoribosyl-dephospho-coA synthase from Archaeoglobus fulgidus
To be Published
|
|
3QSG
 
 | |
5WHM
 
 | | Crystal Structure of IclR Family Transcriptional Regulator from Brucella abortus | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Kim, Y, Wu, R, Tesar, C, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-07-17 | | Release date: | 2017-08-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular control of gene expression byBrucellaBaaR, an IclR-type transcriptional repressor.
J. Biol. Chem., 293, 2018
|
|
3RQZ
 
 | | Crystal structure of metallophosphoesterase from Sphaerobacter thermophilus | | Descriptor: | ACETATE ION, Metallophosphoesterase, ZINC ION | | Authors: | Chang, C, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-28 | | Release date: | 2011-05-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of metallophosphoesterase from Sphaerobacter thermophilus
To be Published
|
|
3ONQ
 
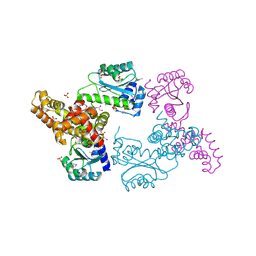 | | Crystal Structure of Regulator of Polyketide Synthase Expression BAD_0249 from Bifidobacterium adolescentis | | Descriptor: | GLYCEROL, Regulator of polyketide synthase expression, SULFATE ION | | Authors: | Kim, Y, Wu, R, Tan, K, Morales, J, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-30 | | Release date: | 2010-09-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Crystal Structure of Regulator of Polyketide Synthase Expression BAD_0249 from Bifidobacterium adolescentis
To be Published
|
|
3QSJ
 
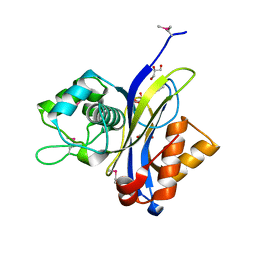 | | Crystal structure of NUDIX hydrolase from Alicyclobacillus acidocaldarius | | Descriptor: | CALCIUM ION, GLYCEROL, NUDIX hydrolase | | Authors: | Michalska, K, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-21 | | Release date: | 2011-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of NUDIX hydrolase from Alicyclobacillus acidocaldarius
To be Published
|
|
3TVA
 
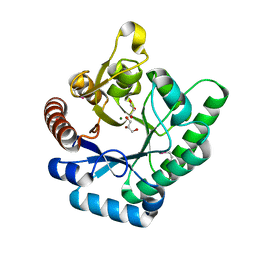 | | Crystal Structure of Xylose isomerase domain protein from Planctomyces limnophilus | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, Y, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-19 | | Release date: | 2011-10-05 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Crystal Structure of Xylose isomerase domain protein from Planctomyces limnophilus
To be Published
|
|
3HF7
 
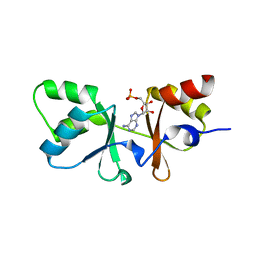 | | The Crystal Structure of a CBS-domain Pair with Bound AMP from Klebsiella pneumoniae to 2.75A | | Descriptor: | ADENOSINE MONOPHOSPHATE, uncharacterized CBS-domain protein | | Authors: | Stein, A.J, Nocek, B, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-11 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Crystal Structure of a CBS-domain Pair with Bound AMP from Klebsiella pneumoniae to 2.75A
To be Published
|
|
3R0V
 
 | | The crystal structure of an alpha/beta hydrolase from Sphaerobacter thermophilus DSM 20745. | | Descriptor: | Alpha/beta hydrolase fold protein, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Tan, K, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-09 | | Release date: | 2011-04-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.383 Å) | | Cite: | The crystal structure of an alpha/beta hydrolase from Sphaerobacter thermophilus DSM 20745.
To be Published
|
|
3RPJ
 
 | | Structure of a curlin genes transcriptional regulator protein from Proteus mirabilis HI4320. | | Descriptor: | 1,2-ETHANEDIOL, Curlin genes transcriptional regulator, SULFATE ION | | Authors: | Cuff, M.E, Wu, R, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-31 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a curlin genes transcriptional regulator protein from Proteus mirabilis HI4320.
TO BE PUBLISHED
|
|
3IVL
 
 | |
4ESY
 
 | | Crystal Structure of the CBS Domain of CBS Domain Containing Membrane Protein from Sphaerobacter thermophilus | | Descriptor: | 1,2-ETHANEDIOL, CBS domain containing membrane protein, CHLORIDE ION | | Authors: | Kim, Y, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-23 | | Release date: | 2012-09-05 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Crystal Structure of the CBS Domain of CBS Domain Containing Membrane Protein from Sphaerobacter thermophilus
To be Published
|
|
