3A4X
 
 | | Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase, GLYCEROL, ... | | Authors: | Tsuji, H, Nishimura, S, Inui, T, Ishikawa, K, Nakamura, T, Uegaki, K. | | Deposit date: | 2009-07-22 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Kinetic and crystallographic analyses of the catalytic domain of chitinase from Pyrococcus furiosus- the role of conserved residues in the active site
Febs J., 277, 2010
|
|
3A4W
 
 | | Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, MAGNESIUM ION, ... | | Authors: | Tsuji, H, Nishimura, S, Inui, T, Ishikawa, K, Nakamura, T, Uegaki, K. | | Deposit date: | 2009-07-22 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and crystallographic analyses of the catalytic domain of chitinase from Pyrococcus furiosus- the role of conserved residues in the active site
Febs J., 277, 2010
|
|
3ALQ
 
 | | Crystal structure of TNF-TNFR2 complex | | Descriptor: | COBALT (II) ION, Tumor necrosis factor, Tumor necrosis factor receptor superfamily member 1B | | Authors: | Mukai, Y, Nakamura, T, Yamagata, Y, Tsutsumi, Y. | | Deposit date: | 2010-08-06 | | Release date: | 2010-11-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Solution of the Structure of the TNF-TNFR2 Complex
Sci.Signal., 3, 2010
|
|
5ZSS
 
 | |
8H3M
 
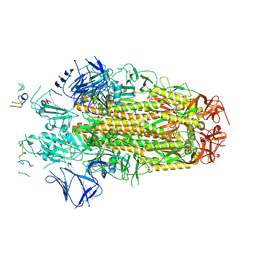 | | Conformation 1 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy chain, Spike glycoprotein | | Authors: | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
8H3N
 
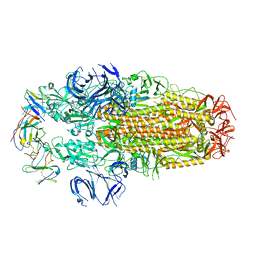 | | Conformation 2 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy-chain, MO1 light chain, ... | | Authors: | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
8Q8J
 
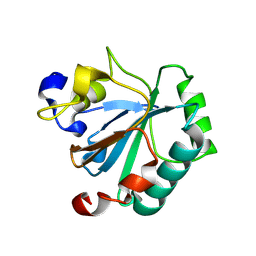 | | Crystal structure of human GPX4-R152H | | Descriptor: | Glutathione peroxidase | | Authors: | Napolitano, V, Mourao, A, Kolonko, M, Bostock, M.J, Sattler, M, Conrad, M, Popowicz, G. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human GPX4-R152H
To Be Published
|
|
8Q8N
 
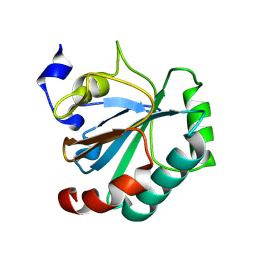 | | Crystal structure of human GPX4-U46C-I129S-L130S | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Bostock, M.J, Mourao, A, Napolitano, V, Sattler, M, Conrad, M, Popowicz, G. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An ultra-rare variant of GPX4 reveals the structural basis to avert neurodegeneration
To Be Published
|
|
3L85
 
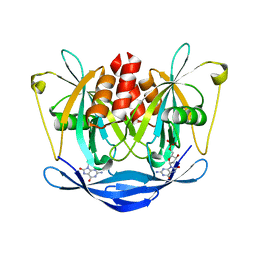 | |
9J1N
 
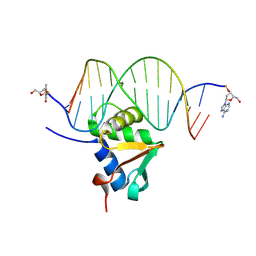 | | Mouse Spi-B Ets domain in complex with DNA containing AGAA sequence | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, DNA (5'-D(*(3D1)P*TP*GP*AP*AP*AP*AP*GP*AP*GP*AP*AP*TP*TP*GP*G)-3'), DNA (5'-D(*(THM)P*CP*CP*AP*AP*TP*TP*CP*TP*CP*TP*TP*TP*TP*CP*A)-3'), ... | | Authors: | Nonaka, Y, Kamitori, S. | | Deposit date: | 2024-08-05 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural analysis of Spi-B DNA-binding Ets domain recognizing 5'-AGAA-3' and 5'-GGAA-3' sequences.
Biochem.Biophys.Res.Commun., 749, 2025
|
|
9J1O
 
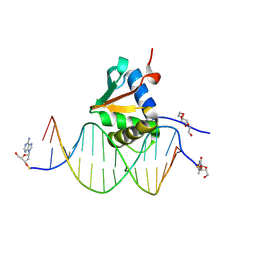 | | Mouse Spi-B Ets domain in complex with DNA containing GGAA sequence | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, DNA (5'-D(*(3D1)P*TP*GP*AP*AP*AP*AP*GP*GP*GP*AP*AP*TP*TP*GP*G)-3'), DNA (5'-D(*(THM)P*CP*CP*AP*AP*TP*TP*CP*CP*CP*TP*TP*TP*TP*CP*A)-3'), ... | | Authors: | Nonaka, Y, Kamitori, S. | | Deposit date: | 2024-08-05 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural analysis of Spi-B DNA-binding Ets domain recognizing 5'-AGAA-3' and 5'-GGAA-3' sequences.
Biochem.Biophys.Res.Commun., 749, 2025
|
|
6D0D
 
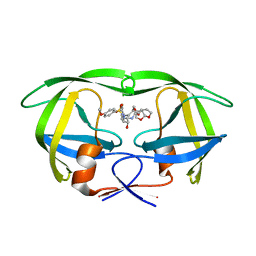 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-087-13 | | Descriptor: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-1-(4-fluorophenyl)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}butan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Novel Central Nervous System (CNS)-Targeting Protease Inhibitors for Drug-Resistant HIV Infection and HIV-Associated CNS Complications.
Antimicrob.Agents Chemother., 63, 2019
|
|
6D0E
 
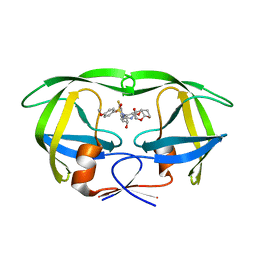 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-084-13 | | Descriptor: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-1-(3,5-difluorophenyl)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}butan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel Central Nervous System (CNS)-Targeting Protease Inhibitors for Drug-Resistant HIV Infection and HIV-Associated CNS Complications.
Antimicrob.Agents Chemother., 63, 2019
|
|
2EAL
 
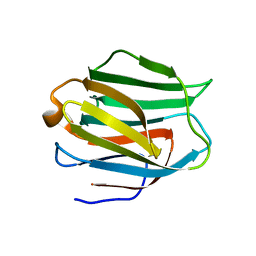 | | Crystal structure of human galectin-9 N-terminal CRD in complex with Forssman pentasaccharide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-beta-D-galactopyranose, Galectin-9 | | Authors: | Nagae, M, Nakamura-Tsuruta, S, Nishi, N, Nakamura, T, Hirabayashi, J, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-09-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of the human galectin-9 N-terminal carbohydrate recognition domain reveals unexpected properties that differ from the mouse orthologue.
J.Mol.Biol., 375, 2008
|
|
2EAK
 
 | | Crystal structure of human galectin-9 N-terminal CRD in complex with lactose | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, GLYCEROL, Galectin-9, ... | | Authors: | Nagae, M, Nakamura-Tsuruta, S, Nishi, N, Nakamura, T, Hirabayashi, J, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-09-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural analysis of the human galectin-9 N-terminal carbohydrate recognition domain reveals unexpected properties that differ from the mouse orthologue.
J.Mol.Biol., 375, 2008
|
|
2RTS
 
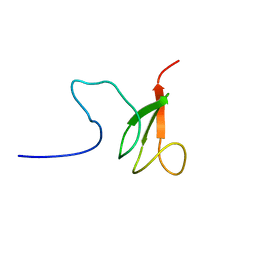 | | Chitin binding domain1 | | Descriptor: | chitinase | | Authors: | Uegaki, K. | | Deposit date: | 2013-08-19 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the chitin-binding domain 1 (ChBD1) of a hyperthermophilic chitinase from Pyrococcus furiosus.
J.Biochem., 155, 2014
|
|
4RKY
 
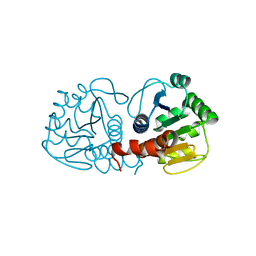 | | Crystal structure of DJ-1 isoform X1 | | Descriptor: | Protein DJ-1 | | Authors: | Liddington, R.C. | | Deposit date: | 2014-10-14 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transnitrosylation from DJ-1 to PTEN attenuates neuronal cell death in parkinson's disease models.
J.Neurosci., 34, 2014
|
|
4RKW
 
 | | Crystal structure of DJ-1 | | Descriptor: | Protein DJ-1 | | Authors: | Liddington, R.C. | | Deposit date: | 2014-10-14 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transnitrosylation from DJ-1 to PTEN attenuates neuronal cell death in parkinson's disease models.
J.Neurosci., 34, 2014
|
|
6L68
 
 | | X-ray structure of human galectin-10 in complex with D-allose | | Descriptor: | Galectin-10, beta-D-allopyranose | | Authors: | Kamitori, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of human galectin-10/monosaccharide complexes demonstrate potential of monosaccharides as effectors in forming Charcot-Leyden crystals.
Biochem.Biophys.Res.Commun., 2020
|
|
6L67
 
 | | X-ray structure of human galectin-10 in complex with D-galactose | | Descriptor: | Galectin-10, beta-D-galactopyranose | | Authors: | Kamitori, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structures of human galectin-10/monosaccharide complexes demonstrate potential of monosaccharides as effectors in forming Charcot-Leyden crystals.
Biochem.Biophys.Res.Commun., 2020
|
|
6L6D
 
 | | X-ray structure of human galectin-10 in complex with D-N-acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, Galectin-10 | | Authors: | Kamitori, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structures of human galectin-10/monosaccharide complexes demonstrate potential of monosaccharides as effectors in forming Charcot-Leyden crystals.
Biochem.Biophys.Res.Commun., 2020
|
|
6L6B
 
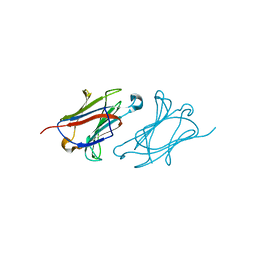 | | X-ray structure of human galectin-10 in complex with L-fucose | | Descriptor: | Galectin-10, beta-L-fucopyranose | | Authors: | Kamitori, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structures of human galectin-10/monosaccharide complexes demonstrate potential of monosaccharides as effectors in forming Charcot-Leyden crystals.
Biochem.Biophys.Res.Commun., 2020
|
|
6L6C
 
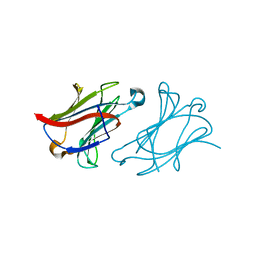 | | X-ray structure of human galectin-10 in complex with D-arabinose | | Descriptor: | Galectin-10, alpha-D-arabinopyranose | | Authors: | Kamitori, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structures of human galectin-10/monosaccharide complexes demonstrate potential of monosaccharides as effectors in forming Charcot-Leyden crystals.
Biochem.Biophys.Res.Commun., 2020
|
|
6L64
 
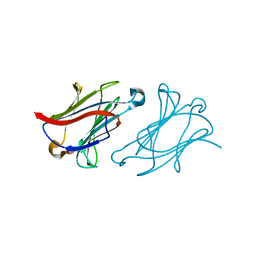 | | X-ray structure of human galectin-10 in complex with D-glucose | | Descriptor: | Galectin-10, beta-D-glucopyranose | | Authors: | Kamitori, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structures of human galectin-10/monosaccharide complexes demonstrate potential of monosaccharides as effectors in forming Charcot-Leyden crystals.
Biochem.Biophys.Res.Commun., 2020
|
|
6L6A
 
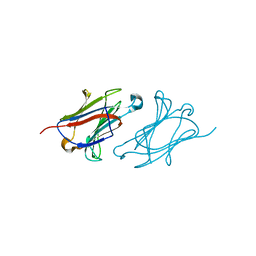 | | X-ray structure of human galectin-10 in complex with D-mannose | | Descriptor: | Galectin-10, beta-D-mannopyranose | | Authors: | Kamitori, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structures of human galectin-10/monosaccharide complexes demonstrate potential of monosaccharides as effectors in forming Charcot-Leyden crystals.
Biochem.Biophys.Res.Commun., 2020
|
|
