2C27
 
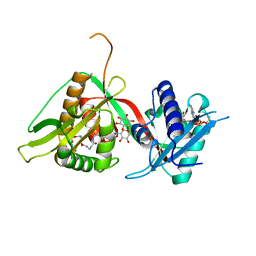 | | The Structure of Mycothiol Synthase in Complex with des- AcetylMycothiol and CoenzymeA. | | Descriptor: | (1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl 2-(L-cysteinylamino)-2-deoxy-alpha-D-glucopyranoside, ACETYL COENZYME *A, COENZYME A, ... | | Authors: | Vetting, M.W, Yu, M, Rendle, P.M, Blanchard, J.S. | | Deposit date: | 2005-09-26 | | Release date: | 2005-12-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Substrate-Induced Conformational Change of Mycobacterium Tuberculosis Mycothiol Synthase.
J.Biol.Chem., 281, 2006
|
|
2BM5
 
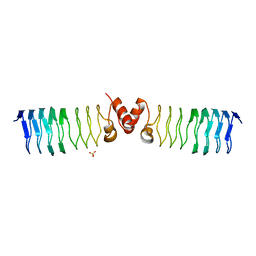 | | The Structure of MfpA (Rv3361c, P21 Crystal form). The Pentapeptide Repeat Protein from Mycobacterium tuberculosis Folds as A Right- handed Quadrilateral Beta-helix. | | Descriptor: | PENTAPEPTIDE REPEAT FAMILY PROTEIN, SULFATE ION | | Authors: | Hegde, S.S, Vetting, M.W, Roderick, S.L, Mitchenall, L.A, Maxwell, A, Takiff, H.E, Blanchard, J.S. | | Deposit date: | 2005-03-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Fluroquinolone Resistance Protein from Mycobacterium Tuberculosis that Mimics DNA
Science, 308, 2005
|
|
2BM6
 
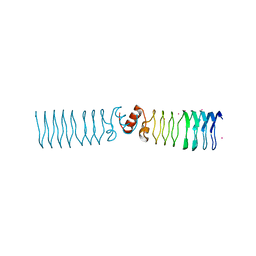 | | The Structure of MfpA (Rv3361c, C2221 Crystal form). The Pentapeptide Repeat Protein from Mycobacterium tuberculosis Folds as A Right- handed Quadrilateral Beta-helix. | | Descriptor: | CESIUM ION, PENTAPEPTIDE REPEAT FAMILY PROTEIN | | Authors: | Hegde, S.S, Vetting, M.W, Roderick, S.L, Mitchenall, L.A, Maxwell, A, Takiff, H.E, Blanchard, J.S. | | Deposit date: | 2005-03-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Fluroquinolone Resistance Protein from Mycobacterium Tuberculosis that Mimics DNA
Science, 308, 2005
|
|
1RYQ
 
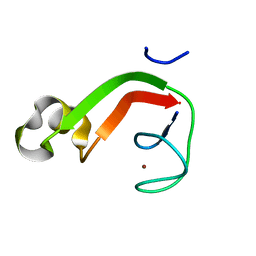 | | Putative DNA-directed RNA polymerase, subunit e'' from Pyrococcus Furiosus Pfu-263306-001 | | Descriptor: | DNA-directed RNA polymerase, subunit e'', ZINC ION | | Authors: | Liu, Z.-J, Chen, L, Tempel, W, Shah, A, Arendall III, W.B, Rose, J.P, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Lee, H.S, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Parameter-space screening: a powerful tool for high-throughput crystal structure determination.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2GKE
 
 | | Crystal structure of diaminopimelate epimerase in complex with an irreversible inhibitor LL-AziDAP | | Descriptor: | (2S,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, ACETIC ACID, Diaminopimelate epimerase, ... | | Authors: | Pillai, B, Cherney, M.M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C, James, M.N. | | Deposit date: | 2006-04-01 | | Release date: | 2006-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into stereochemical inversion by diaminopimelate epimerase: An antibacterial drug target.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1S7F
 
 | | RimL- Ribosomal L7/L12 alpha-N-protein acetyltransferase crystal form I (apo) | | Descriptor: | CHLORIDE ION, MALONIC ACID, acetyl transferase | | Authors: | Vetting, M.W, de Carvalho, L.P, Roderick, S.L, Blanchard, J.S. | | Deposit date: | 2004-01-29 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel dimeric structure of the RimL Nalpha-acetyltransferase from Salmonella typhimurium.
J.Biol.Chem., 280, 2005
|
|
1Q5N
 
 | |
1S7L
 
 | | RimL- Ribosomal L7/L12 alpha-N-protein acetyltransferase in complex with Coenzyme A (CoA-Cys134 Disulfide) | | Descriptor: | COENZYME A, SULFATE ION, acetyl transferase | | Authors: | Vetting, M.W, de Carvalho, L.P, Roderick, S.L, Blanchard, J.S. | | Deposit date: | 2004-01-29 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel dimeric structure of the RimL Nalpha-acetyltransferase from Salmonella typhimurium.
J.Biol.Chem., 280, 2005
|
|
2CNM
 
 | | RimI - Ribosomal S18 N-alpha-protein acetyltransferase in complex with a bisubstrate inhibitor (Cterm-Arg-Arg-Phe-Tyr-Arg-Ala-N-alpha- acetyl-S-CoA). | | Descriptor: | 30S RIBOSOMAL PROTEIN S18, COENZYME A, MODIFICATION OF 30S RIBOSOMAL SUBUNIT PROTEIN S18 | | Authors: | Vetting, M.W, Yu, M, Bareich, D.C, Blanchard, J.S. | | Deposit date: | 2006-05-22 | | Release date: | 2007-05-22 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Rimi from Salmonella Typhimurium Lt2, the Gnat Responsible for N{Alpha}- Acetylation of Ribosomal Protein S18.
Protein Sci., 17, 2008
|
|
2BUE
 
 | | Structure of AAC(6')-Ib in complex with Ribostamycin and Coenzyme A. | | Descriptor: | AAC(6')-IB, CALCIUM ION, COENZYME A, ... | | Authors: | Vetting, M.W, Park, C.H, Hedge, S.S, Hooper, D.C, Blanchard, J.S. | | Deposit date: | 2008-03-20 | | Release date: | 2008-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanistic and Structural Analysis of Aminoglycoside N-Acetyltransferase Aac(6')-Ib and its Bifunctional, Fluoroquinolone-Active Aac(6')-Ib-Cr Variant.
Biochemistry, 47, 2008
|
|
1S7K
 
 | |
2BM4
 
 | | The Structure of MfpA (Rv3361c, C2 Crystal form). The Pentapeptide Repeat Protein from Mycobacterium tuberculosis Folds as A Right- handed Quadrilateral Beta-helix. | | Descriptor: | PENTAPEPTIDE REPEAT FAMILY PROTEIN | | Authors: | Hegde, S.S, Vetting, M.W, Roderick, S.L, Mitchenall, L.A, Maxwell, A, Takiff, H.E, Blanchard, J.S. | | Deposit date: | 2005-03-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Fluroquinolone Resistance Protein from Mycobacterium Tuberculosis that Mimics DNA
Science, 308, 2005
|
|
2GKJ
 
 | | Crystal structure of diaminopimelate epimerase in complex with an irreversible inhibitor DL-AZIDAP | | Descriptor: | (2R,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, ACETIC ACID, Diaminopimelate epimerase | | Authors: | Pillai, B, Cherney, M.M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C, James, M.N. | | Deposit date: | 2006-04-02 | | Release date: | 2006-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into stereochemical inversion by diaminopimelate epimerase: An antibacterial drug target.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1S7N
 
 | | Ribosomal L7/L12 alpha-N-protein acetyltransferase in complex with Coenzyme A (CoA free sulfhydryl) | | Descriptor: | COENZYME A, acetyl transferase | | Authors: | Vetting, M.W, de Carvalho, L.P, Roderick, S.L, Blanchard, J.S. | | Deposit date: | 2004-01-29 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel dimeric structure of the RimL Nalpha-acetyltransferase from Salmonella typhimurium.
J.Biol.Chem., 280, 2005
|
|
2CNT
 
 | | RimI - Ribosomal S18 N-alpha-protein acetyltransferase in complex with CoenzymeA. | | Descriptor: | COENZYME A, GLYCEROL, MODIFICATION OF 30S RIBOSOMAL SUBUNIT PROTEIN S18, ... | | Authors: | Vetting, M.W, Bareich, D.C, Yu, M, Blanchard, J.S. | | Deposit date: | 2006-05-23 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Rimi from Salmonella Typhimurium Lt2, the Gnat Responsible for N{Alpha}- Acetylation of Ribosomal Protein S18.
Protein Sci., 17, 2008
|
|
4OKZ
 
 | | Selinadiene Synthase in complex with dihydrofarnesyl pyrophosphate | | Descriptor: | (3S,6E)-3,7,11-trimethyldodeca-6,10-dien-1-yl trihydrogen diphosphate, MAGNESIUM ION, Terpene synthase metal-binding domain-containing protein | | Authors: | Baer, P, Rabe, P, Fischer, K, Citron, C.A, Klapschinski, T, Groll, M, Dickschat, J.S. | | Deposit date: | 2014-01-23 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Induced-fit mechanism in class i terpene cyclases.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4OKM
 
 | | Selinadiene Synthase apo and in complex with diphosphate | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE, Terpene synthase metal-binding domain-containing protein | | Authors: | Baer, P, Rabe, P, Fischer, K, Citron, C.A, Klapschinski, T, Groll, M, Dickschat, J.S. | | Deposit date: | 2014-01-22 | | Release date: | 2014-06-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induced-fit mechanism in class i terpene cyclases.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2J8K
 
 | | Structure of the fusion of NP275 and NP276, pentapeptide repeat proteins from Nostoc punctiforme | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NP275-NP276, SULFATE ION | | Authors: | Vetting, M.W, Hegde, S.S, Hazleton, K.Z, Blanchard, J.S. | | Deposit date: | 2006-10-25 | | Release date: | 2006-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Characterization of the Fusion of Two Pentapeptide Repeat Proteins, Np275 and Np276, from Nostoc Punctiforme: Resurrection of an Ancestral Protein.
Protein Sci., 16, 2007
|
|
2J8I
 
 | | Structure of NP275, a pentapeptide repeat protein from Nostoc punctiforme | | Descriptor: | CHLORIDE ION, NP275 | | Authors: | Vetting, M.W, Hegde, S.S, Hazleton, K.Z, Blanchard, J.S. | | Deposit date: | 2006-10-25 | | Release date: | 2006-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of the Fusion of Two Pentapeptide Repeat Proteins, Np275 and Np276, from Nostoc Punctiforme: Resurrection of an Ancestral Protein.
Protein Sci., 16, 2007
|
|
2Q9J
 
 | | Crystal structure of the C217S mutant of diaminopimelate epimerase | | Descriptor: | 1,2-ETHANEDIOL, Diaminopimelate epimerase, SULFATE ION | | Authors: | Pillai, B, Cherney, M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C. | | Deposit date: | 2007-06-12 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamics of catalysis revealed from the crystal structures of mutants of diaminopimelate epimerase.
Biochem.Biophys.Res.Commun., 363, 2007
|
|
2PR2
 
 | |
2Q9H
 
 | | Crystal structure of the C73S mutant of diaminopimelate epimerase | | Descriptor: | ACETIC ACID, Diaminopimelate epimerase, L(+)-TARTARIC ACID | | Authors: | Pillai, B, Cherney, M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C, James, M.N.G. | | Deposit date: | 2007-06-12 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamics of catalysis revealed from the crystal structures of mutants of diaminopimelate epimerase.
Biochem.Biophys.Res.Commun., 363, 2007
|
|
2VBQ
 
 | | Structure of AAC(6')-Iy in complex with bisubstrate analog CoA-S- monomethyl-acetylneamine. | | Descriptor: | (3R,9Z)-17-[(2R,3S,4R,5R,6R)-5-amino-6-{[(1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl]oxy}-3,4-dihydroxytetrahydro-2H-pyran-2-yl]-3-hydroxy-2,2-dimethyl-4,8,15-trioxo-12-thia-5,9,16-triazaheptadec-9-en-1-yl [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, AMINOGLYCOSIDE 6'-N-ACETYLTRANSFERASE, GLYCEROL, ... | | Authors: | Vetting, M.W, Magalhaes, M.L, Freiburger, L, Gao, F, Auclair, K, Blanchard, J.S. | | Deposit date: | 2007-09-14 | | Release date: | 2008-01-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and Structural Analysis of Bisubstrate Inhibition of the Salmonella Enterica Aminoglycoside 6'-N-Acetyltransferase.
Biochemistry, 47, 2008
|
|
3C4Q
 
 | | Structure of the retaining glycosyltransferase MshA : The first step in mycothiol biosynthesis. Organism : Corynebacterium glutamicum- Complex with UDP | | Descriptor: | MAGNESIUM ION, Predicted glycosyltransferases, SULFATE ION, ... | | Authors: | Vetting, M.W, Frantom, P.A, Blanchard, J.S. | | Deposit date: | 2008-01-30 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Enzymatic Analysis of MshA from Corynebacterium glutamicum: SUBSTRATE-ASSISTED CATALYSIS
J.Biol.Chem., 283, 2008
|
|
3C48
 
 | |
