5N4C
 
 | | Prolyl oligopeptidase B from Galerina marginata bound to 35mer hydrolysis and macrocyclization substrate - S577A mutant | | Descriptor: | Alpha-amanitin proprotein, GLYCEROL, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
5NC4
 
 | | Crystal structure of the ferric enterobactin receptor (PfeA) in complex with protochelin from Pseudomonas aeruginosa | | Descriptor: | FE (III) ION, Ferric enterobactin receptor, ~{N}-[(5~{S})-5-[[2,3-bis(oxidanyl)phenyl]carbonylamino]-6-[4-[[2,3-bis(oxidanyl)phenyl]carbonylamino]butylamino]-6-oxidanylidene-hexyl]-2,3-bis(oxidanyl)benzamide | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2017-03-03 | | Release date: | 2018-03-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The complex of ferric-enterobactin with its transporter from Pseudomonas aeruginosa suggests a two-site model.
Nat Commun, 10, 2019
|
|
5N4E
 
 | | Prolyl oligopeptidase B from Galerina marginata bound to 35mer hydrolysis and macrocyclization substrate - H698A mutant | | Descriptor: | Alpha-amanitin proprotein, GLYCEROL, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
5NJN
 
 | | Roll out the beta-barrel: structure and mechanism of Pac13, a unique nucleoside dehydratase | | Descriptor: | Putative cupin_2 domain-containing isomerase | | Authors: | Michailidou, F, Bent, A.F, Naismith, J.H, Goss, R.J.M. | | Deposit date: | 2017-03-29 | | Release date: | 2018-03-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pac13 is a Small, Monomeric Dehydratase that Mediates the Formation of the 3'-Deoxy Nucleoside of Pacidamycins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5NC3
 
 | |
5NC8
 
 | | Shewanella denitrificans Kef CTD in AMP bound form | | Descriptor: | ADENOSINE MONOPHOSPHATE, Potassium efflux system protein | | Authors: | Pliotas, C, Naismith, J.H. | | Deposit date: | 2017-03-03 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Adenosine Monophosphate Binding Stabilizes the KTN Domain of the Shewanella denitrificans Kef Potassium Efflux System.
Biochemistry, 56, 2017
|
|
5O3V
 
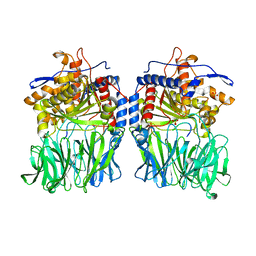 | | Structural characterization of the fast and promiscuous macrocyclase from plant - PCY1-S562A bound to Presegetalin B1 | | Descriptor: | MAGNESIUM ION, Peptide cyclase 1, Putative presegetalin B1, ... | | Authors: | Ludewig, H, Czekster, C.M, Bent, A.F, Naismith, J.H. | | Deposit date: | 2017-05-25 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Characterization of the Fast and Promiscuous Macrocyclase from Plant PCY1 Enables the Use of Simple Substrates.
ACS Chem. Biol., 13, 2018
|
|
5O3U
 
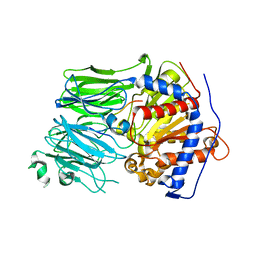 | | Structural characterization of the fast and promiscuous macrocyclase from plant - PCY1-S562A bound to Presegetalin F1 | | Descriptor: | Peptide cyclase 1, Putative presegetalin F1 | | Authors: | Ludewig, H, Czekster, C.M, Bent, A.F, Naismith, J.H. | | Deposit date: | 2017-05-25 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Characterization of the Fast and Promiscuous Macrocyclase from Plant PCY1 Enables the Use of Simple Substrates.
ACS Chem. Biol., 13, 2018
|
|
5NR2
 
 | |
5N0R
 
 | |
5N4I
 
 | |
1RTV
 
 | | RmlC (dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase) crystal structure from Pseudomonas aeruginosa, apo structure | | Descriptor: | S,R MESO-TARTARIC ACID, dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Dong, C.J, Naismith, J.H. | | Deposit date: | 2003-12-10 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RmlC (dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase) crystal structure from Pseudomonas aeruginosa, apo structure
TO BE PUBLISHED
|
|
9G97
 
 | | Lipid III flippase WzxE with NB10 nanobody in outward-facing conformation at 0.9688 A | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, CHLORIDE ION, Lipid III flippase, ... | | Authors: | Le Bas, A, El Omari, K, Lee, M, Naismith, J.H. | | Deposit date: | 2024-07-24 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of WzxE the lipid III flippase for Enterobacterial Common Antigen polysaccharide.
Open Biology, 15, 2025
|
|
9G9N
 
 | |
9G95
 
 | | Lipid III flippase WzxE with NB10 nanobody in outward-facing conformation at 2.7552 A | | Descriptor: | CHLORIDE ION, Lipid III flippase, N-OCTANE, ... | | Authors: | Le Bas, A, El Omari, K, Lee, M, Naismith, J.H. | | Deposit date: | 2024-07-24 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of WzxE the lipid III flippase for Enterobacterial Common Antigen polysaccharide.
Open Biology, 15, 2025
|
|
9G9P
 
 | |
9G9M
 
 | |
9G9O
 
 | |
2VH3
 
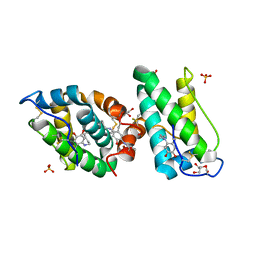 | | ranasmurfin | | Descriptor: | GLYCEROL, RANASMURFIN, SULFATE ION, ... | | Authors: | Oke, M, Ching, R.T, Carter, L.G, Johnson, K.A, Liu, H, McMahon, S.A, Bloch Junior, C, Botting, C.H, Walsh, M.A, Latiff, A.A, Kennedy, M.W, Cooper, A, Naismith, J.H. | | Deposit date: | 2007-11-17 | | Release date: | 2007-12-04 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Unusual Chromophore and Cross-Links in Ranasmurfin: A Blue Protein from the Foam Nests of a Tropical Frog.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
1SCS
 
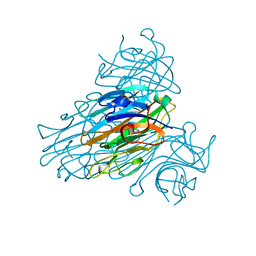 | | HIGH-RESOLUTION STRUCTURES OF SINGLE-METAL-SUBSTITUTED CONCANAVALIN A: THE CO,CA-PROTEIN AT 1.6 ANGSTROMS AND THE NI,CA-PROTEIN AT 2.0 ANGSTROMS | | Descriptor: | CALCIUM ION, COBALT (II) ION, CONCANAVALIN A | | Authors: | Emmerich, C, Helliwell, J.R, Redshaw, M, Naismith, J.H, Harrop, S.J, Raftery, J, Kalb, A.J, Yariv, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 1993-12-06 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structures of single-metal-substituted concanavalin A: the Co,Ca-protein at 1.6 A and the Ni,Ca-protein at 2.0 A.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1SCR
 
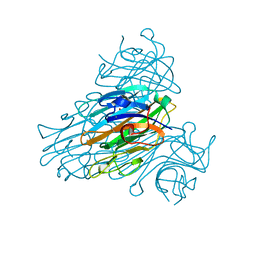 | | HIGH-RESOLUTION STRUCTURES OF SINGLE-METAL-SUBSTITUTED CONCANAVALIN A: THE CO,CA-PROTEIN AT 1.6 ANGSTROMS AND THE NI,CA-PROTEIN AT 2.0 ANGSTROMS | | Descriptor: | CALCIUM ION, CONCANAVALIN A, NICKEL (II) ION | | Authors: | Emmerich, C, Helliwell, J.R, Redshaw, M, Naismith, J.H, Harrop, S.J, Raftery, J, Kalb, A.J, Yariv, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 1993-12-06 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of single-metal-substituted concanavalin A: the Co,Ca-protein at 1.6 A and the Ni,Ca-protein at 2.0 A.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
5AIU
 
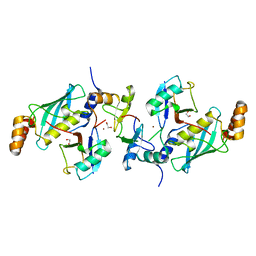 | | A complex of RNF4-RING domain, Ubc13-Ub (isopeptide crosslink) | | Descriptor: | 1,2-ETHANEDIOL, E3 UBIQUITIN-PROTEIN LIGASE RNF4, POLYUBIQUITIN-C, ... | | Authors: | Branigan, E, Naismith, J.H. | | Deposit date: | 2015-02-17 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis for the Ring Catalyzed Synthesis of K63 Linked Ubiquitin Chains
Nat.Struct.Mol.Biol., 22, 2015
|
|
5AIT
 
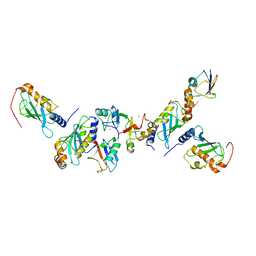 | | A complex of of RNF4-RING domain, UbeV2, Ubc13-Ub (isopeptide crosslink) | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE RNF4, POLYUBIQUITIN-C, UBIQUITIN-CONJUGATING ENZYME E2 N, ... | | Authors: | Branigan, E, Naismith, J.H. | | Deposit date: | 2015-02-17 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis for the Ring Catalyzed Synthesis of K63 Linked Ubiquitin Chains
Nat.Struct.Mol.Biol., 22, 2015
|
|
1O7I
 
 | |
1OI6
 
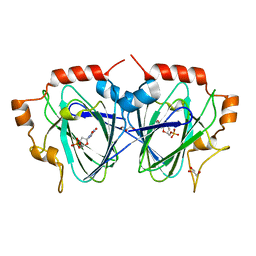 | | Structure determination of the TMP-complex of EvaD | | Descriptor: | GLYCEROL, PCZA361.16, THYMIDINE-5'-PHOSPHATE | | Authors: | Merkel, A.B, Naismith, J.H. | | Deposit date: | 2003-06-09 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Position of a Key Tyrosine in Dtdp-4-Keto-6-Deoxy-D-Glucose-5-Epimerase (Evad) Alters the Substrate Profile for This Rmlc-Like Enzyme
J.Biol.Chem., 279, 2004
|
|
