7YLW
 
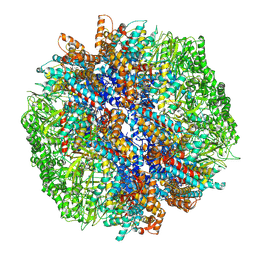 | | yeast TRiC-plp2-tubulin complex at S3 closed TRiC state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
7YLX
 
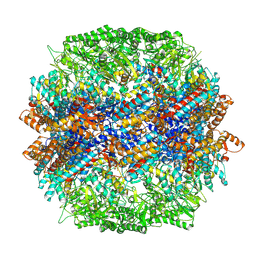 | | yeast TRiC-plp2-actin complex at S4 closed TRiC state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
7YLY
 
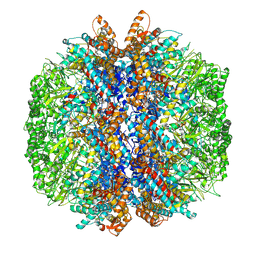 | | yeast TRiC-plp2 complex at S5 closed TRiC state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
8WBV
 
 | |
8WBU
 
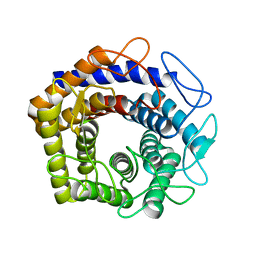 | |
9B97
 
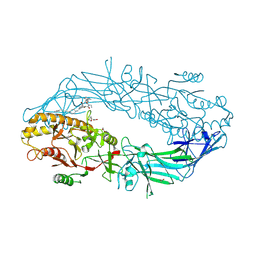 | | Crystal structure of the human PAD2 protein bound to small molecule | | Descriptor: | (1P)-N~3'~-[(2S)-3-cyclohexyl-1-(methylamino)-1-oxopropan-2-yl]-N~3~,N~3~-diethyl-4-fluoro-5'-{[4-(4-phenylbutyl)piperazin-1-yl]methyl}[1,1'-biphenyl]-3,3'-dicarboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Byrnes, L.J, Vajdos, F. | | Deposit date: | 2024-04-01 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery, Characterization, and Structure of a Cell Active PAD2 Inhibitor Acting through a Novel Allosteric Mechanism.
Acs Chem.Biol., 19, 2024
|
|
9B96
 
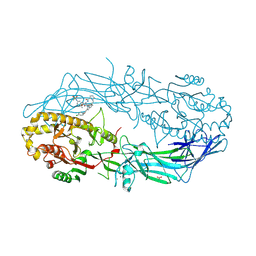 | | Crystal structure of the human PAD2 protein bound to inhibitor | | Descriptor: | 1-({(2P)-1-{(1R)-1-(2-bromophenyl)-3-[5-(methanesulfonamido)-2-methylanilino]-3-oxopropyl}-2-[3-(4-chlorophenoxy)phenyl]-1H-1,3-benzimidazol-6-yl}methyl)-N-methyl-D-prolinamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Byrnes, L.J, Vajdos, F. | | Deposit date: | 2024-04-01 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery, Characterization, and Structure of a Cell Active PAD2 Inhibitor Acting through a Novel Allosteric Mechanism.
Acs Chem.Biol., 19, 2024
|
|
9B98
 
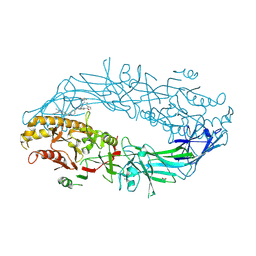 | | Crystal structure of the human PAD2 protein bound to small molecule | | Descriptor: | (5P)-N,N-diethyl-2-fluoro-5-(2-[({1-[2-(methylamino)-2-oxoethyl]cyclohexyl}methyl)amino]-6-{methyl[1-(2-methyl-1-phenyl-1H-1,3-benzimidazole-5-carbonyl)piperidin-4-yl]amino}pyrimidin-4-yl)benzamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Byrnes, L.J, Vajdos, F. | | Deposit date: | 2024-04-01 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Discovery, Characterization, and Structure of a Cell Active PAD2 Inhibitor Acting through a Novel Allosteric Mechanism.
Acs Chem.Biol., 19, 2024
|
|
8XBS
 
 | | C. elegans apo-SID1 structure | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gong, D.S. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Structural basis for double-stranded RNA recognition by SID1.
Nucleic Acids Res., 52, 2024
|
|
8XC1
 
 | | C. elegans SID1 in complex with dsRNA | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gong, D.S. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Structural basis for double-stranded RNA recognition by SID1.
Nucleic Acids Res., 52, 2024
|
|
4EWS
 
 | |
7C8W
 
 | | Structure of sybody MR17 in complex with the SARS-CoV-2 S receptor-binding domain (RBD) | | Descriptor: | GLYCEROL, Spike protein S1, Synthetic nanobody MR17, ... | | Authors: | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7C8V
 
 | | Structure of sybody SR4 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7CAN
 
 | | Structure of sybody MR17-K99Y in complex with the SARS-CoV-2 S Receptor-binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-09 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7WVP
 
 | |
7WVQ
 
 | |
7D5G
 
 | |
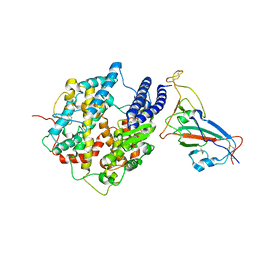
7WK5
 
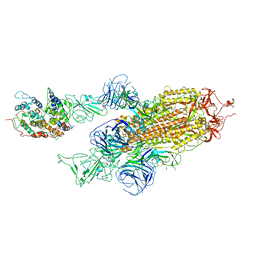 | | Cryo-EM structure of Omicron S-ACE2, C2 state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Han, W.Y, Wang, Y.F. | | Deposit date: | 2022-01-08 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Molecular basis of SARS-CoV-2 Omicron variant receptor engagement and antibody evasion and neutralization
Biorxiv, 2022
|
|
7WK4
 
 | |
7WK6
 
 | |
7Y63
 
 | | ApoSIDT2-pH7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 2, ... | | Authors: | Gong, D.S. | | Deposit date: | 2022-06-18 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural insight into the human SID1 transmembrane family member 2 reveals its lipid hydrolytic activity.
Nat Commun, 14, 2023
|
|
7Y68
 
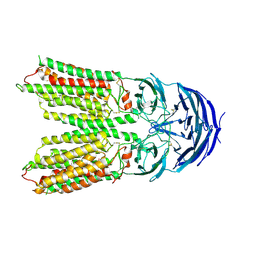 | | SIDT2-pH5.5 plus miRNA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 2, ... | | Authors: | Gong, D.S. | | Deposit date: | 2022-06-18 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural insight into the human SID1 transmembrane family member 2 reveals its lipid hydrolytic activity.
Nat Commun, 14, 2023
|
|
7Y69
 
 | | ApoSIDT2-pH5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 2, ... | | Authors: | Gong, D.S. | | Deposit date: | 2022-06-18 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural insight into the human SID1 transmembrane family member 2 reveals its lipid hydrolytic activity.
Nat Commun, 14, 2023
|
|
8IVZ
 
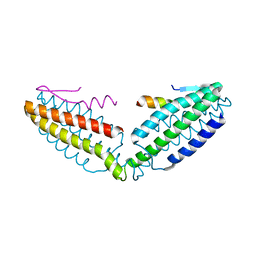 | | Crystal structure of talin R7 in complex with KANK1 KN motif | | Descriptor: | KN motif and ankyrin repeat domains 1, Talin-1 | | Authors: | Xu, Y, Li, K, Wei, Z, Cong, Y. | | Deposit date: | 2023-03-29 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
7D2S
 
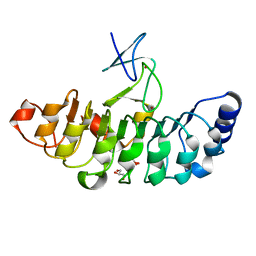 | | Crystal structure of Rsu1/PINCH1_LIM5C complex | | Descriptor: | GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, Ras suppressor protein 1, ... | | Authors: | Yang, H, Wei, Z, Cong, Y. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
