1GFA
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GFK
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GFU
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GF9
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GFJ
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1INU
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GAZ
 
 | | Crystal Structure of Mutant Human Lysozyme Substituted at the Surface Positions | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues at turns in the conformational stability and folding of human lysozyme.
Biochemistry, 39, 2000
|
|
1GB5
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions.
Biochemistry, 39, 2000
|
|
1GFE
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GBO
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions.
Biochemistry, 39, 2000
|
|
1GFV
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
6M4E
 
 | | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Uehara, R, Iwamoto, R, Aoki, S, Yoshizawa, T, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis.
Protein Sci., 29, 2020
|
|
6M55
 
 | | Crystal structure of the E496A mutant of HsBglA in complex with 4-galactosyllactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase-like enzyme, ... | | Authors: | Uehara, R, Iwamoto, R, Aoki, S, Yoshizawa, T, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2020-03-10 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis.
Protein Sci., 29, 2020
|
|
6M4F
 
 | | Crystal structure of the E496A mutant of HsBglA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase-like enzyme, ... | | Authors: | Uehara, R, Iwamoto, R, Aoki, S, Yoshizawa, T, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis.
Protein Sci., 29, 2020
|
|
5AVH
 
 | | The 0.90 angstrom structure (I222) of glucose isomerase crystallized in high-strength agarose hydrogel | | Descriptor: | Xylose isomerase | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, N, Sazaki, G, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
5AVN
 
 | | The 1.03 angstrom structure (P212121) of glucose isomerase crystallized in high-strength agarose hydrogel | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, N, Sazaki, G, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-23 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
5AVG
 
 | | The 0.95 angstrom structure of thaumatin crystallized in high-strength agarose hydrogel | | Descriptor: | Thaumatin-1 | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, M, Sazaki, G, Hirose, M, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
5AVD
 
 | | The 0.86 angstrom structure of elastase crystallized in high-strength agarose hydrogel | | Descriptor: | Chymotrypsin-like elastase family member 1, SULFATE ION | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, M, Sazaki, G, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
5X7K
 
 | |
1IOC
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME, EAEA-I56T | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Goda, S, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-02-27 | | Release date: | 2002-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Elongation in a beta-structure promotes amyloid-like fibril formation of human lysozyme.
J.Biochem., 132, 2002
|
|
2E1P
 
 | | Crystal structure of pro-Tk-subtilisin | | Descriptor: | CALCIUM ION, Tk-subtilisin | | Authors: | Tanaka, S, Saito, K, Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2006-10-27 | | Release date: | 2007-01-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of unautoprocessed precursor of subtilisin from a hyperthermophilic archaeon: evidence for Ca2+-induced folding
J.Biol.Chem., 282, 2007
|
|
3WR7
 
 | | Crystal Structure of Spermidine Acetyltransferase from Escherichia coli | | Descriptor: | COENZYME A, SPERMIDINE, Spermidine N1-acetyltransferase | | Authors: | Sugiyama, S, Ishikawa, S, Tomitori, S, Niiyama, M, Hirose, M, Miyazaki, Y, Higashi, K, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Kashiwagi, K, Igarashi, K, Matsumura, H. | | Deposit date: | 2014-02-20 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism underlying promiscuous polyamine recognition by spermidine acetyltransferase
Int.J.Biochem.Cell Biol., 76, 2016
|
|
2D0C
 
 | | Crystal structure of Bst-RNase HIII in complex with Mn2+ | | Descriptor: | MANGANESE (II) ION, ribonuclease HIII | | Authors: | Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2005-07-31 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and structure-based mutational analyses of RNase HIII from Bacillus stearothermophilus: a new type 2 RNase H with TBP-like substrate-binding domain at the N terminus
J.Mol.Biol., 356, 2006
|
|
2D0B
 
 | | Crystal structure of Bst-RNase HIII in complex with Mg2+ | | Descriptor: | MAGNESIUM ION, ribonuclease HIII | | Authors: | Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2005-07-31 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and structure-based mutational analyses of RNase HIII from Bacillus stearothermophilus: a new type 2 RNase H with TBP-like substrate-binding domain at the N terminus
J.Mol.Biol., 356, 2006
|
|
3VND
 
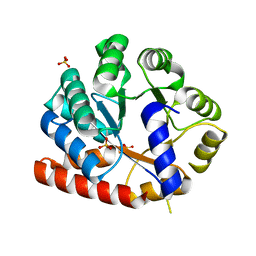 | | Crystal structure of tryptophan synthase alpha-subunit from the psychrophile Shewanella frigidimarina K14-2 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, SULFATE ION, Tryptophan synthase alpha chain | | Authors: | Mitsuya, D, Tanaka, S, Matsumura, H, Takano, K, Urano, N, Ishida, M. | | Deposit date: | 2012-01-12 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Strategy for cold adaptation of the tryptophan synthase alpha subunit from the psychrophile Shewanella frigidimarina K14-2: crystal structure and physicochemical properties
J.Biochem., 155, 2014
|
|
