8B24
 
 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 3600-seconds post reaction initiation with Na+ | | Descriptor: | DIPHOSPHATE, K(+)-stimulated pyrophosphate-energized sodium pump, MAGNESIUM ION, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (4.53 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
8CD8
 
 | | Ulilysin - C269A with AEBSF complex | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CALCIUM ION, GLY-SER-SER, ... | | Authors: | Rodriguez-Banqueri, A, Eckhard, U, Gomis-Ruth, F.X. | | Deposit date: | 2023-01-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into latency of the metallopeptidase ulilysin (lysargiNase) and its unexpected inhibition by a sulfonyl-fluoride inhibitor of serine peptidases.
Dalton Trans, 52, 2023
|
|
8CEM
 
 | | Structure of bovine native C3, re-refinement | | Descriptor: | Complement C3 alpha chain, Complement C3 beta chain, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Andersen, G.R, Fredslund, F. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of bovine complement component 3 reveals the basis for thioester function.
J.Mol.Biol., 361, 2006
|
|
8BRX
 
 | | Escherichia coli methionyl-tRNA synthetase mutant L13C,I297C complexed with beta-3-methionine | | Descriptor: | (3R)-3-amino-5-(methylsulfanyl)pentanoic acid, CITRIC ACID, Methionine--tRNA ligase, ... | | Authors: | Schmitt, E, Mechulam, Y, Nigro, G, Opuu, V, Lazennec-Schurdevin, C, Simonson, T. | | Deposit date: | 2022-11-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Redesigning methionyl-tRNA synthetase for beta-methionine activity with adaptive landscape flattening and experiments.
Protein Sci., 32, 2023
|
|
8C3P
 
 | |
8C3O
 
 | | Crystal structure of autotaxin gamma and compound MEY-003 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5,7-bis(oxidanyl)-2-(1-pentylindol-3-yl)chromen-4-one, 7alpha-hydroxycholesterol, ... | | Authors: | Eymery, M.C, McCarthy, A.A. | | Deposit date: | 2022-12-27 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery of potent chromone-based autotaxin inhibitors inspired by cannabinoids.
Eur.J.Med.Chem., 263, 2023
|
|
8C7R
 
 | | Crystal structure of rat autotaxin and compound MEY-003 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5,7-bis(oxidanyl)-2-(1-pentylindol-3-yl)chromen-4-one, 7alpha-hydroxycholesterol, ... | | Authors: | Eymery, M.C, McCarthy, A.A. | | Deposit date: | 2023-01-17 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Discovery of potent chromone-based autotaxin inhibitors inspired by cannabinoids.
Eur.J.Med.Chem., 263, 2023
|
|
8C4W
 
 | | Crystal structure of rat autotaxin and compound MEY-002 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5,7-bis(oxidanyl)-2-[1-(phenylmethyl)indol-3-yl]chromen-4-one, 7alpha-hydroxycholesterol, ... | | Authors: | Eymery, M.C, McCarthy, A.A. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of potent chromone-based autotaxin inhibitors inspired by cannabinoids.
Eur.J.Med.Chem., 263, 2023
|
|
8C3T
 
 | |
8D86
 
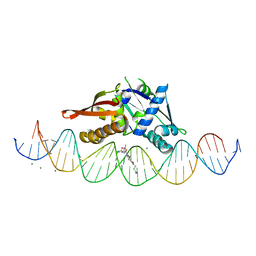 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence co-crystallized with a guest small molecule, netropsin. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3'), DNA (5'-D(A*GP*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Snow, C.D. | | Deposit date: | 2022-06-07 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
8D8M
 
 | |
8ASO
 
 | | Nickel(II) bound to a non-canonical quadruplex | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3'), NICKEL (II) ION | | Authors: | Lambert, M.C, Hall, J.P. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Identifying metal-DNA binding sites, what is the best method to get transition metals into a crystal system?
To Be Published
|
|
8ASM
 
 | | Cobalt(II) bound to a non-canonical quadruplex | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3') | | Authors: | Lambert, M.C, Hall, J.P. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identifying metal-DNA binding sites, what is the best method to get transition metals into a crystal system?
To Be Published
|
|
4FE1
 
 | | Improving the Accuracy of Macromolecular Structure Refinement at 7 A Resolution | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Fromme, R, Adams, P.D, Fromme, P, Levitt, M, Schroeder, G.F, Brunger, A.T. | | Deposit date: | 2012-05-29 | | Release date: | 2012-08-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.9228 Å) | | Cite: | Improving the accuracy of macromolecular structure refinement at 7 A resolution.
Structure, 20, 2012
|
|
6P0J
 
 | | Crystal structure of GDP-bound human RalA | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Ral-A | | Authors: | Bum-Erdene, K, Gonzalez-Gutierrez, G, Liu, D, Meroueh, S.O. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PHJ
 
 | |
6PHO
 
 | |
6PHK
 
 | |
6PHP
 
 | |
6PQ7
 
 | |
6PHN
 
 | |
6PHI
 
 | |
6PHM
 
 | |
6PHL
 
 | |
6PHQ
 
 | |
