4DRX
 
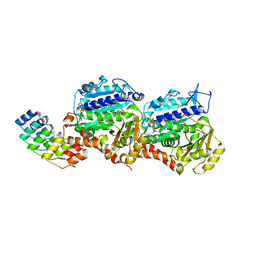 | | GTP-Tubulin in complex with a DARPIN | | Descriptor: | Designed ankyrin repeat protein (DARPIN) D1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pecqueur, L, Gigant, B, Knossow, M. | | Deposit date: | 2012-02-17 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A designed ankyrin repeat protein selected to bind to tubulin caps the microtubule plus end.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5JS8
 
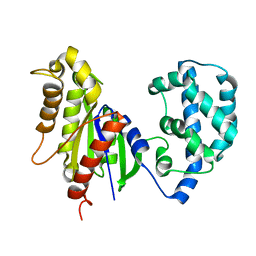 | | Structural Model of a Protein alpha subunit in complex with GDP obtained with SAXS and NMR residual couplings | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1 | | Authors: | Goricanec, D, Stehle, R, Grigoriu, S, Wagner, G, Hagn, F. | | Deposit date: | 2016-05-07 | | Release date: | 2016-06-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational dynamics of a G-protein alpha subunit is tightly regulated by nucleotide binding.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JS7
 
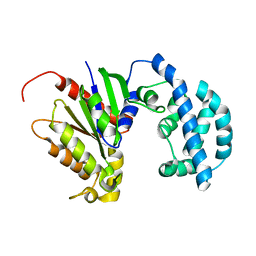 | | Structural model of a apo G-protein alpha subunit determined with NMR residual dipolar couplings and SAXS | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1 | | Authors: | Goricanec, D, Stehle, R, Grigoriu, S, Wagner, G, Hagn, F. | | Deposit date: | 2016-05-07 | | Release date: | 2016-06-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational dynamics of a G-protein alpha subunit is tightly regulated by nucleotide binding.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1N3N
 
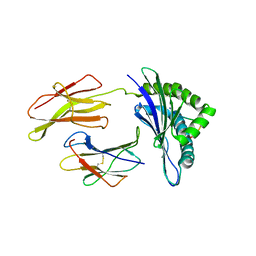 | | Crystal structure of a mycobacterial hsp60 epitope with the murine class I MHC molecule H-2Db | | Descriptor: | BETA-2-MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B ALPHA CHAIN, ... | | Authors: | Ciatto, C, Capitani, G, Tissot, A.C, Pecorari, F, Pluckthun, A, Grutter, M.G. | | Deposit date: | 2002-10-29 | | Release date: | 2003-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of mycobacterial
and murine hsp60 epitopes in
complex with the class I MHC
molecule H-2D(b)
FEBS Lett., 543, 2003
|
|
7PDH
 
 | |
7PD4
 
 | |
7PDE
 
 | |
7PD8
 
 | |
7PDD
 
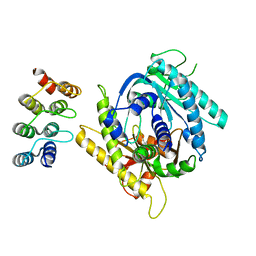 | |
7PDG
 
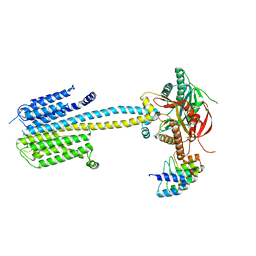 | |
7PDF
 
 | |
5LE4
 
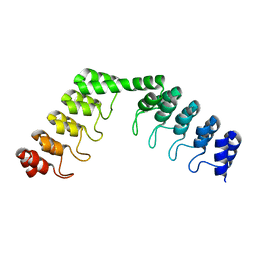 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_11_D12 | | Descriptor: | DD_D12_11_D12 | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LEA
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_Off7_12_3G124 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DD_Off7_12_3G124 | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LED
 
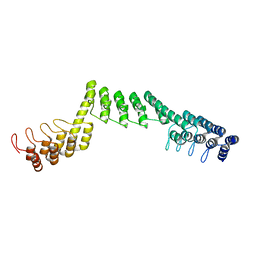 | | Crystal structure of DARPin-DARPin rigid fusion, variant DDD_D12_12_D12_12_D12 | | Descriptor: | DDD_D12_12_D12_12_D12 | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LEM
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_Off7_11_3G124 in complex with Maltose-binding Protein and Green Fluorescent Protein | | Descriptor: | DD_Off7_11_3G124, Green fluorescent protein, Maltose-binding periplasmic protein | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5MA8
 
 | | GFP-binding DARPin 3G124nc | | Descriptor: | GA-binding protein subunit beta-1, Green fluorescent protein | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MA6
 
 | | GFP-binding DARPin 3G124nc | | Descriptor: | 1,2-ETHANEDIOL, 3G124nc, Green fluorescent protein, ... | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MAK
 
 | | GFP-binding DARPin fusion gc_R7 | | Descriptor: | CITRIC ACID, Green fluorescent protein, R7 | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MA4
 
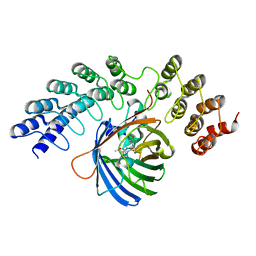 | | GFP-binding DARPin fusion gc_K7 | | Descriptor: | Green fluorescent protein, K7 | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MAD
 
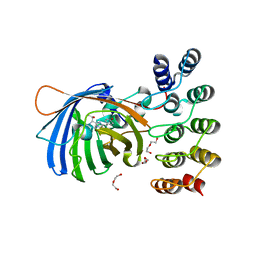 | | GFP-binding DARPin 3G61 | | Descriptor: | 3G61, DI(HYDROXYETHYL)ETHER, Green fluorescent protein, ... | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
6S9R
 
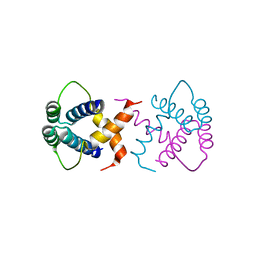 | | Crystal structure of SSDP from D. melanogaster | | Descriptor: | Sequence-specific single-stranded DNA-binding protein, isoform A | | Authors: | Renko, M, Bienz, M. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rotational symmetry of the structured Chip/LDB-SSDP core module of the Wnt enhanceosome.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5MA3
 
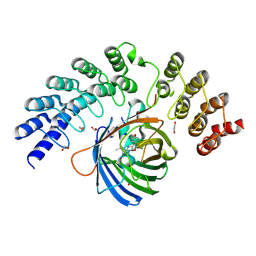 | | GFP-binding DARPin fusion gc_R11 | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, R11 | | Authors: | Hansen, S, Stueber, J, Ernst, P, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MA5
 
 | | GFP-binding DARPin fusion gc_K11 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Green fluorescent protein, ... | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MA9
 
 | | GFP-binding DARPin fusion gc_R11 | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, R11 | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5LW1
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_232_11_D12 in complex JNK1a1 and JIP1 peptide | | Descriptor: | ADENOSINE, C-Jun-amino-terminal kinase-interacting protein 1, DD_232_11_D12, ... | | Authors: | Wu, Y, Batyuk, A, Mittl, P.R, Honegger, A, Plueckthun, A. | | Deposit date: | 2016-09-15 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for the Selective Inhibition of c-Jun N-Terminal Kinase 1 Determined by Rigid DARPin-DARPin Fusions.
J.Mol.Biol., 430, 2018
|
|
