6YTR
 
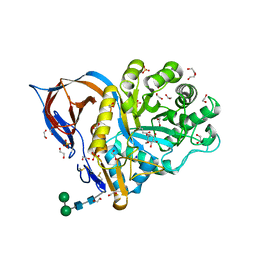 | | Structure of recombinant human beta-glucocerebrosidase in complex with cyclophellitol aziridine inhibitor | | Descriptor: | (1~{R},2~{S},3~{S},4~{S},5~{R},6~{R})-5-azanyl-6-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2020-04-24 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, Synthesis and Structural Analysis of Glucocerebrosidase Imaging Agents.
Chemistry, 27, 2021
|
|
6YUT
 
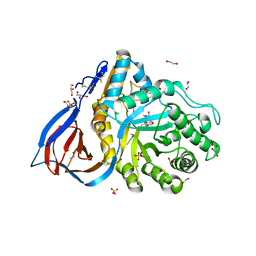 | | Structure of recombinant human beta-glucocerebrosidase in complex with N-acyl functionalised cyclophellitol aziridine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2020-04-27 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Design, Synthesis and Structural Analysis of Glucocerebrosidase Imaging Agents.
Chemistry, 27, 2021
|
|
6Z39
 
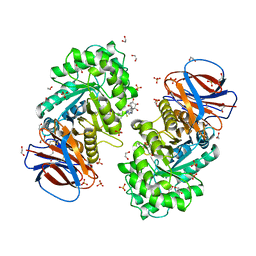 | | Structure of recombinant human beta-glucocerebrosidase in complex with BODIPY functionalised epoxide activity based probe | | Descriptor: | (1~{S},2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[4-[4-[(12~{R})-2,2-bis(fluoranyl)-4,6,10,12-tetramethyl-1,3-diaza-2$l^{4}-boratricyclo[7.3.0.0^{3,7}]dodeca-4,6,9-trien-8-yl]butyl]-1,2,3-triazol-1-yl]methyl]-7-oxabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2020-05-19 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, Synthesis and Structural Analysis of Glucocerebrosidase Imaging Agents.
Chemistry, 27, 2021
|
|
6Z3I
 
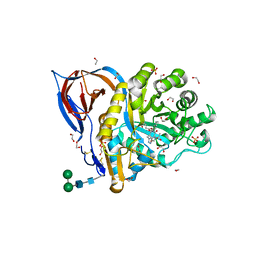 | | Structure of recombinant beta-glucocerebrosidase in complex with bifunctional cyclophellitol aziridine activity based probe | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosomal acid glucosylceramidase, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2020-05-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis and Structural Analysis of Glucocerebrosidase Imaging Agents.
Chemistry, 27, 2021
|
|
6ZPV
 
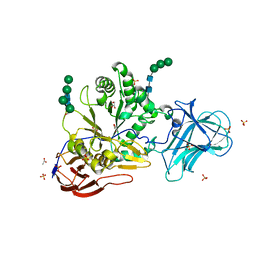 | | Structure of Unliganded MgGH51 a-L-Arabinofuranosidase Crystal Type 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZQ0
 
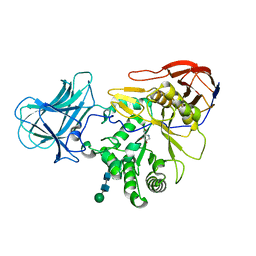 | | Structure of a-l-AraAZI-Bound MgGH51 a-L-Arabinofuranosidase Crystal Type 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MgGH51, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZPW
 
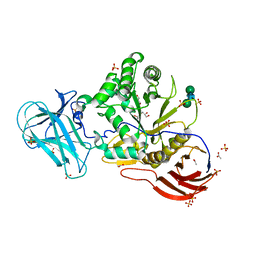 | | Structure of Unliganded MgGH51 a-L-Arabinofuranosidase Crystal Type 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CHLORIDE ION, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.329 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZPZ
 
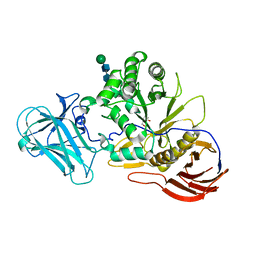 | | Structure of a-l-AraCS-Bound MgGH51 a-L-Arabinofuranosidase Crystal Type 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MgGH51, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZQ1
 
 | | Structure of AraDNJ-Bound MgGH51 a-L-Arabinofuranosidase Crystal Type 1 | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-L-ARABINITOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZPS
 
 | |
6ZPX
 
 | | Structure of Unliganded MgGH51 a-L-Arabinofuranosidase Crystal Type 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MgGH51, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZPY
 
 | | Structure of Arabinose-Bound MgGH51 a-L-Arabinofuranosidase Crystal Type 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MgGH51, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6SXR
 
 | | E221Q mutant of GH54 a-l-arabinofuranosidase soaked with 4-nitrophenyl a-l-arabinofuranoside | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGregor, N.G.S, Davies, G.J, Nin-Hill, A, Rovira, C. | | Deposit date: | 2019-09-26 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Rational Design of Mechanism-Based Inhibitors and Activity-Based Probes for the Identification of Retaining alpha-l-Arabinofuranosidases.
J.Am.Chem.Soc., 142, 2020
|
|
7ODJ
 
 | | Exo-mannosidase from Cellvibrio mixtus bound to N-alkyl mannocyclophellitol aziridine | | Descriptor: | (1R,2S,3R,4S,5R,6R)-5-(8-azidooctylamino)-6-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, ACETATE ION, Man5A, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2021-04-29 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Synthesis of broad-specificity activity-based probes for exo -beta-mannosidases.
Org.Biomol.Chem., 20, 2022
|
|
7NWV
 
 | | Structure of recombinant human beta-glucocerebrosidase in complex with BODIPY Tagged Cyclophellitol activity based probe | | Descriptor: | (1~{S},2~{R},3~{R},4~{S},5~{S})-4-[[4-[4-[2,2-bis(fluoranyl)-4,6,10,12-tetramethyl-3-aza-1-azonia-2-boranuidatricyclo[7.3.0.0^{3,7}]dodeca-1(12),4,6,8,10-pentaen-8-yl]butyl]-1,2,3-triazol-1-yl]methyl]cyclohexane-1,2,3,5-tetrol, 1,2-ETHANEDIOL, 1-deoxy-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2021-03-17 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fluorescence polarisation activity-based protein profiling for the identification of deoxynojirimycin-type inhibitors selective for lysosomal retaining alpha- and beta-glucosidases.
Chem Sci, 14, 2023
|
|
7OMI
 
 | | Bs164 in complex with mannocyclophellitol epoxide | | Descriptor: | (1R,2S,4S,5R)-6-(hydroxymethyl)cyclohexane-1,2,3,4,5-pentol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Armstrong, Z, Mcgregor, N, Davies, G. | | Deposit date: | 2021-05-24 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Synthesis of broad-specificity activity-based probes for exo -beta-mannosidases.
Org.Biomol.Chem., 20, 2022
|
|
7P4C
 
 | | Crystal Structure of Agd31B, alpha-transglucosylase in Glycoside Hydrolase Family 31, in complex with noncovalent Cyclophellitol Sulfamidate probe KK131 | | Descriptor: | (3aR,4S,5S,6R,7R,7aS)-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3a,4,5,6,7,7a-hexahydro-3H-benzo[d][1,2,3]oxathiazole-4,5,6-triol, 1,2-ETHANEDIOL, OXALATE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | 1,6- epi-Cyclophellitol Cyclosulfamidate Is a Bona Fide Lysosomal alpha-Glucosidase Stabilizer for the Treatment of Pompe Disease.
J.Am.Chem.Soc., 144, 2022
|
|
7P4D
 
 | | Crystal Structure of Agd31B, alpha-transglucosylase in Glycoside Hydrolase Family 31, in complex with covalent Cyclophellitol Sulfamidate probe KK130 | | Descriptor: | 1,2-ETHANEDIOL, OXALATE ION, Oligosaccharide 4-alpha-D-glucosyltransferase, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1,6- epi-Cyclophellitol Cyclosulfamidate Is a Bona Fide Lysosomal alpha-Glucosidase Stabilizer for the Treatment of Pompe Disease.
J.Am.Chem.Soc., 144, 2022
|
|
5L5Z
 
 | |
5L6C
 
 | |
5L5B
 
 | |
5L5W
 
 | |
5L54
 
 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 16 | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-~{N}-[(2~{S},3~{S},4~{R})-4-methyl-3,5-bis(oxidanyl)-1-phenyl-pentan-2-yl]-2-[[(2~{R})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
5L5T
 
 | | Yeast 20S proteasome with human beta5i (1-138; V31M) and human beta6 (97-111; 118-133) in complex with epoxyketone inhibitor 16 | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-~{N}-[(2~{S},3~{S},4~{R})-4-methyl-3,5-bis(oxidanyl)-1-phenyl-pentan-2-yl]-2-[[(2~{R})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
5L68
 
 | |
