8TMC
 
 | |
8TMF
 
 | |
8TMH
 
 | |
8TMP
 
 | |
8TMI
 
 | |
8TMJ
 
 | |
8TMO
 
 | |
8TMK
 
 | |
8TMM
 
 | |
8TMD
 
 | |
8TMN
 
 | |
8TML
 
 | |
8TME
 
 | |
8TMQ
 
 | |
5PTP
 
 | |
9MXV
 
 | | Human Vault Cage in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Major vault protein | | Authors: | Lodwick, J.E, Zhao, M. | | Deposit date: | 2025-01-21 | | Release date: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural Insights into the Roles of PARP4 and NAD + in the Human Vault Cage.
Biorxiv, 2024
|
|
9MXH
 
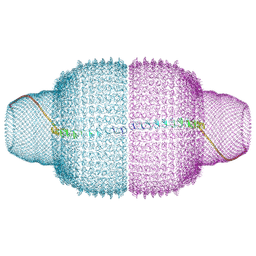 | | Human Vault Cage in complex with NAD+ | | Descriptor: | Major vault protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lodwick, J.E, Zhao, M. | | Deposit date: | 2025-01-20 | | Release date: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural Insights into the Roles of PARP4 and NAD + in the Human Vault Cage.
Biorxiv, 2024
|
|
1NAG
 
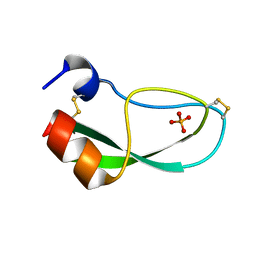 | | CREVICE-FORMING MUTANTS IN THE RIGID CORE OF BOVINE PANCREATIC TRYPSIN INHIBITOR: CRYSTAL STRUCTURES OF F22A, Y23A, N43G, AND F45A | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR, PHOSPHATE ION | | Authors: | Danishefsky, A.T, Wlodawer, A, Kim, K.-S, Tao, F, Woodward, C. | | Deposit date: | 1992-08-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crevice-forming mutants in the rigid core of bovine pancreatic trypsin inhibitor: crystal structures of F22A, Y23A, N43G, and F45A.
Protein Sci., 2, 1993
|
|
8PTI
 
 | | CRYSTAL STRUCTURE OF A Y35G MUTANT OF BOVINE PANCREATIC TRYPSIN INHIBITOR | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR | | Authors: | Housset, D, Kim, K.-S, Fuchs, J, Woodward, C, Wlodawer, A. | | Deposit date: | 1990-12-17 | | Release date: | 1991-04-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a Y35G mutant of bovine pancreatic trypsin inhibitor.
J.Mol.Biol., 220, 1991
|
|
5CWT
 
 | |
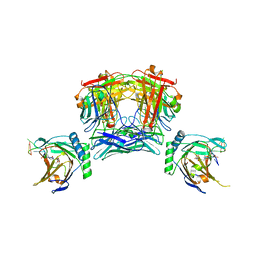
8JNK
 
 | |
8JNR
 
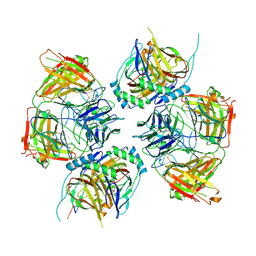 | |
9MJB
 
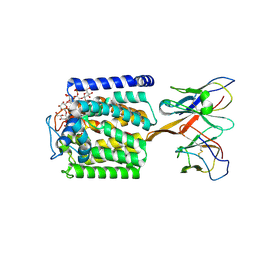 | | Product-Bound mannosyltransferase PimE in complex with Fab | | Descriptor: | (2S)-1-{[(S)-{[(1S,2R,3R,4S,5S,6R)-2-[(6-O-hexadecanoyl-beta-L-gulopyranosyl)oxy]-3,4,5-trihydroxy-6-{[beta-D-mannopyranosyl-(1->2)-alpha-D-mannopyranosyl-(1->6)-beta-D-mannopyranosyl-(1->6)-alpha-D-mannopyranosyl]oxy}cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}-3-(hexadecanoyloxy)propan-2-yl 10-methyloctadecanoate, Fab_E6 heavy chain, Fab_E6 light chain, ... | | Authors: | Liu, Y, Mancia, F. | | Deposit date: | 2024-12-14 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Mechanistic studies of mycobacterial glycolipid biosynthesis by the mannosyltransferase PimE.
Nat Commun, 16, 2025
|
|
3PJS
 
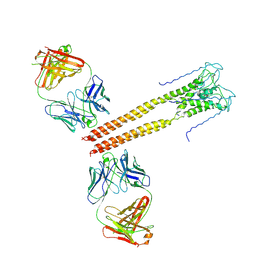 | | Mechanism of Activation Gating in the Full-Length KcsA K+ Channel | | Descriptor: | FAB heavy chain, FAB light chain, Voltage-gated potassium channel | | Authors: | Uysal, S, Cuello, L.G, Kossiakoff, A, Perozo, E. | | Deposit date: | 2010-11-10 | | Release date: | 2011-07-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Mechanism of activation gating in the full-length KcsA K+ channel.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7NIU
 
 | |
