1SZQ
 
 | | Crystal Structure of 2-methylcitrate dehydratase | | Descriptor: | 2-methylcitrate dehydratase | | Authors: | Rajashankar, K.R, Kniewel, R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-04-06 | | Release date: | 2004-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of 2-methylcitrate dehydratase
To be Published
|
|
1Z2I
 
 | |
2EUI
 
 | |
2ESR
 
 | | conserved hypothetical protein- streptococcus pyogenes | | Descriptor: | Methyltransferase, alpha-D-glucopyranose | | Authors: | Jiang, J, Min, T, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-10-26 | | Release date: | 2006-02-07 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of hypothetical protein of Streptococcus Pygenes
To be Published
|
|
1T5J
 
 | |
1TZZ
 
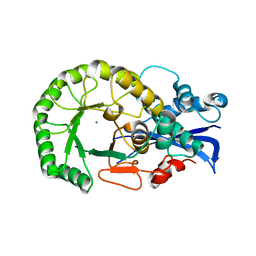 | | Crystal structure of the protein L1841, unknown member of enolase superfamily from Bradyrhizobium japonicum | | Descriptor: | Hypothetical protein L1841, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-12 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the protein L1841, unknown member of enolase superfamily from Bradyrhizobium japonicum
To be Published
|
|
2G6T
 
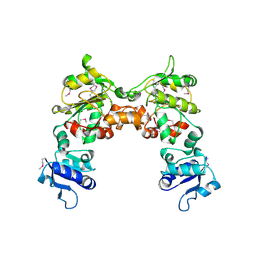 | |
2NV5
 
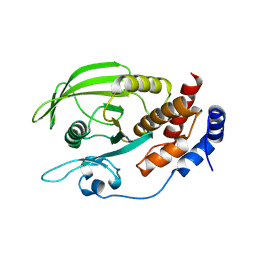 | | Crystal structure of a C-terminal phosphatase domain of Rattus norvegicus ortholog of human protein tyrosine phosphatase, receptor type, D (PTPRD) | | Descriptor: | PTPRD, PHOSPHATASE | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Iizuka, M, Xu, W, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-10 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2AFA
 
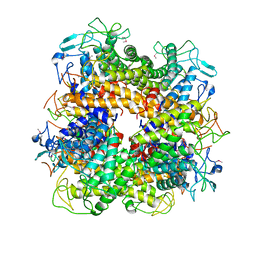 | |
2A1F
 
 | |
1OMI
 
 | | Crystal structure of PrfA,the transcriptional regulator in Listeria monocytogenes | | Descriptor: | GLYCEROL, Listeriolysin regulatory protein | | Authors: | Thirumuruhan, R, Rajashankar, K, Fedorov, A.A, Dodatko, T, Chance, M.R, Cossart, P, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-02-25 | | Release date: | 2003-03-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of PrfA, the transcriptional regulator in Listeria monocytogenes
To be Published
|
|
1P1L
 
 | |
2HSI
 
 | |
2OYC
 
 | | Crystal structure of human pyridoxal phosphate phosphatase | | Descriptor: | Pyridoxal phosphate phosphatase, SODIUM ION, TUNGSTATE(VI)ION | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-21 | | Release date: | 2007-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2P69
 
 | | Crystal Structure of Human Pyridoxal Phosphate Phosphatase with PLP | | Descriptor: | CALCIUM ION, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate phosphatase | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2P8E
 
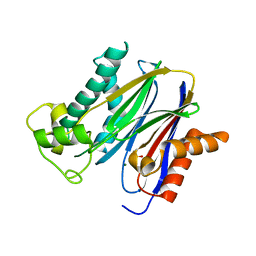 | | Crystal structure of the serine/threonine phosphatase domain of human PPM1B | | Descriptor: | MAGNESIUM ION, PPM1B beta isoform variant 6 | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Lau, C, Xu, W, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-22 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
1Q2Y
 
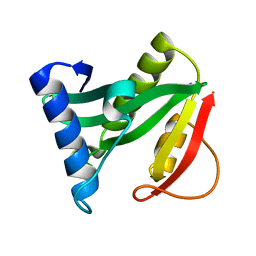 | | Crystal structure of the protein YJCF from Bacillus subtilis: a member of the GCN5-related N-acetyltransferase superfamily fold | | Descriptor: | similar to hypothetical proteins | | Authors: | Fedorov, A.A, Ramagopal, U.A, Fedorov, E.V, Thirumuruhan, R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-07-27 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the protein YJCF from Bacillus subtilis: a member of the GCN5-related N-acetyltransferase superfamily
To be Published
|
|
1PUJ
 
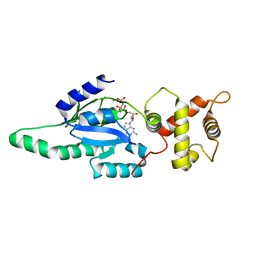 | | Structure of B. subtilis YlqF GTPase | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, conserved hypothetical protein ylqF | | Authors: | Kniewel, R, Buglino, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-24 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the YlqF GTPase from B. subtilis
To be Published
|
|
1PUG
 
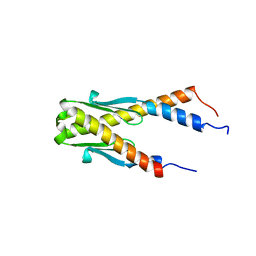 | | Structure of E. coli Ybab | | Descriptor: | Hypothetical UPF0133 protein ybaB | | Authors: | Kniewel, R, Buglino, J, Chadna, T, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-24 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of E. coli Ybab
To be Published
|
|
2PBN
 
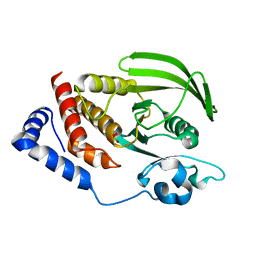 | | Crystal structure of the human tyrosine receptor phosphate gamma | | Descriptor: | Receptor-type tyrosine-protein phosphatase gamma, SULFATE ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Reyes, C, Pelletier, L, Jin, X, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-28 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
1Q6W
 
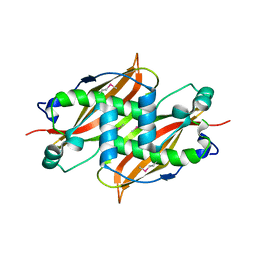 | | X-Ray structure of Monoamine oxidase regulatory protein from Archaeoglobus fulgius | | Descriptor: | monoamine oxidase regulatory protein, putative | | Authors: | Fedorov, A.A, Fedorov, E.V, Thirumuruhan, R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | X-ray structure of monoamine oxidase regulatory protein from Archaeoglobus fulgidus
To be Published
|
|
1RV9
 
 | |
1RI6
 
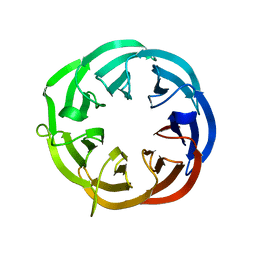 | | Structure of a putative isomerase from E. coli | | Descriptor: | putative isomerase ybhE | | Authors: | Lima, C.D, Kniewel, R, Solorzano, V, Wu, J, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-16 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a putative 7-bladed propeller isomerase
To be Published
|
|
1RW0
 
 | |
1SEF
 
 | |
