8D6Z
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV91-27 | | Descriptor: | Neutralizing antibody COV91-27 heavy chain, Neutralizing antibody COV91-27 light chain, Spike protein S2 fusion peptide | | Authors: | Lee, C.C.D, Lin, T.H, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
9JK4
 
 | | putative E2P of TMEM94 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum magnesium-transporting P-type ATPase | | Authors: | Gong, D.S. | | Deposit date: | 2024-09-15 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural analysis of full-length human transmembrane protein 94 argues against its classification as a P-type Mg 2+ ATPase.
Cell Discov, 11, 2025
|
|
9JK3
 
 | | putative MgE2P of TMEM94 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum magnesium-transporting P-type ATPase | | Authors: | Gong, D.S. | | Deposit date: | 2024-09-14 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural analysis of full-length human transmembrane protein 94 argues against its classification as a P-type Mg 2+ ATPase.
Cell Discov, 11, 2025
|
|
9JJN
 
 | | putative MgE1-ATP of human TMEM94 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum magnesium-transporting P-type ATPase | | Authors: | Gong, D.S. | | Deposit date: | 2024-09-14 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structural analysis of full-length human transmembrane protein 94 argues against its classification as a P-type Mg 2+ ATPase.
Cell Discov, 11, 2025
|
|
9JJO
 
 | | putative MgE1P-ADP of human TMEM94 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum magnesium-transporting P-type ATPase | | Authors: | Gong, D.S. | | Deposit date: | 2024-09-14 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural analysis of full-length human transmembrane protein 94 argues against its classification as a P-type Mg 2+ ATPase.
Cell Discov, 11, 2025
|
|
9JK5
 
 | | putative MgE1-ATP-H of TMEM94 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum magnesium-transporting P-type ATPase | | Authors: | Gong, D.S. | | Deposit date: | 2024-09-15 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural analysis of full-length human transmembrane protein 94 argues against its classification as a P-type Mg 2+ ATPase.
Cell Discov, 11, 2025
|
|
9JJK
 
 | | putative MgE1 of human TMEM94 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum magnesium-transporting P-type ATPase | | Authors: | Gong, D.S. | | Deposit date: | 2024-09-14 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural analysis of full-length human transmembrane protein 94 argues against its classification as a P-type Mg 2+ ATPase.
Cell Discov, 11, 2025
|
|
8DAO
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV44-79 | | Descriptor: | COV44-79 heavy chain constant domain, COV44-79 heavy chain variable domain, COV44-79 light chain constant domain, ... | | Authors: | Lin, T.H, Lee, C.C.D, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
8DTX
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV89-22 | | Descriptor: | COV89-22 heavy chain, COV89-22 light chain, Spike protein S2' stem helix peptide | | Authors: | Lin, T.H, Lee, C.C.D, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTR
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV30-14 | | Descriptor: | COV30-14 heavy chain, COV30-14 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTT
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV93-03 | | Descriptor: | COV93-03 heavy chain, COV93-03 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
7DF4
 
 | | SARS-CoV-2 S-ACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Cong, X, Yao, C. | | Deposit date: | 2020-11-06 | | Release date: | 2020-12-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7DF3
 
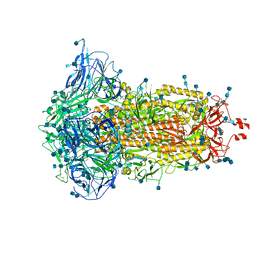 | | SARS-CoV-2 S trimer, S-closed | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cong, X, Yao, C. | | Deposit date: | 2020-11-06 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
2OBD
 
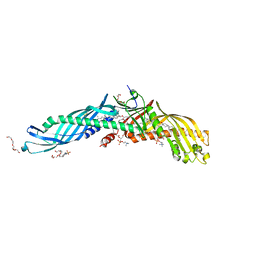 | | Crystal Structure of Cholesteryl Ester Transfer Protein | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Qiu, X. | | Deposit date: | 2006-12-18 | | Release date: | 2007-01-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules.
Nat.Struct.Mol.Biol., 14, 2007
|
|
5WBP
 
 | |
6ZRU
 
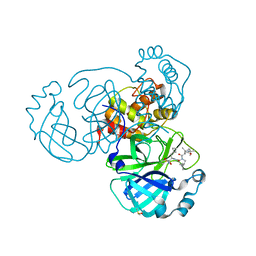 | | Crystal structure of SARS CoV2 main protease in complex with inhibitor Boceprevir | | Descriptor: | DIMETHYL SULFOXIDE, Main Protease, boceprevir (bound form) | | Authors: | Oerlemans, R, Wang, W, Lunev, S, Domling, A, Groves, M.R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Repurposing the HCV NS3-4A protease drug boceprevir as COVID-19 therapeutics.
Rsc Med Chem, 12, 2020
|
|
5WBM
 
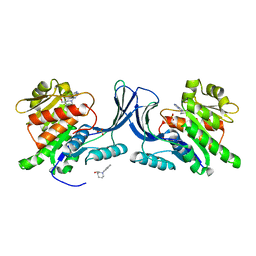 | |
5WBR
 
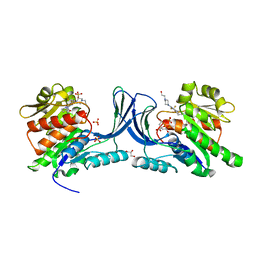 | | Structure of human Ketohexokinase complexed with hits from fragment screening | | Descriptor: | 6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-[(3S)-3-(hydroxymethyl)piperidin-1-yl]-4-(trifluoromethyl)pyridine-3-carbonitrile, CITRIC ACID, GLYCEROL, ... | | Authors: | Pandit, J. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of Fragment-Derived Small Molecules for in Vivo Inhibition of Ketohexokinase (KHK).
J. Med. Chem., 60, 2017
|
|
5WBQ
 
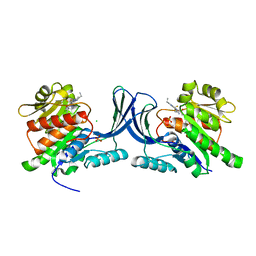 | | Structure of human Ketohexokinase complexed with hits from fragment screening | | Descriptor: | 2-ethyl-7-[(3S)-3-hydroxy-3-methylpyrrolidin-1-yl]-5-(trifluoromethyl)-1H-pyrrolo[3,2-b]pyridine-6-carbonitrile, CHLORIDE ION, Ketohexokinase, ... | | Authors: | Pandit, J. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Fragment-Derived Small Molecules for in Vivo Inhibition of Ketohexokinase (KHK).
J. Med. Chem., 60, 2017
|
|
6ZRT
 
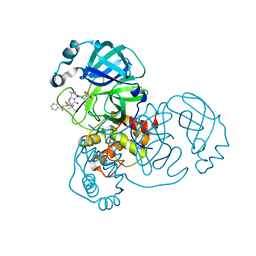 | | Crystal structure of SARS CoV2 main protease in complex with inhibitor Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, DIMETHYL SULFOXIDE, Main Protease | | Authors: | Oerlemans, R, Wang, W, Lunev, S, Domling, A, Groves, M.R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Repurposing the HCV NS3-4A protease drug boceprevir as COVID-19 therapeutics.
Rsc Med Chem, 12, 2020
|
|
5WBO
 
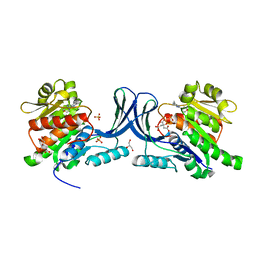 | |
5WBZ
 
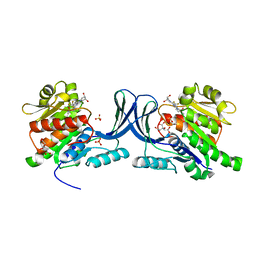 | | Structure of human Ketohexokinase complexed with hits from fragment screening | | Descriptor: | 6-[(3S,4S)-3,4-dihydroxypyrrolidin-1-yl]-2-[(3S)-3-hydroxy-3-methylpyrrolidin-1-yl]-4-(trifluoromethyl)pyridine-3-carbonitrile, CITRIC ACID, Ketohexokinase, ... | | Authors: | Pandit, J. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Fragment-Derived Small Molecules for in Vivo Inhibition of Ketohexokinase (KHK).
J. Med. Chem., 60, 2017
|
|
3J0G
 
 | |
7YLU
 
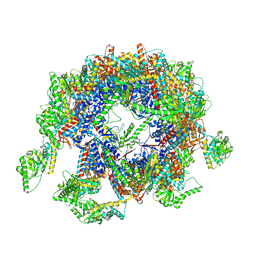 | | yeast TRiC-plp2-substrate complex at S1 TRiC-NPP state | | Descriptor: | Phosducin-like protein 2, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
7YLV
 
 | | yeast TRiC-plp2-substrate complex at S2 ATP binding state | | Descriptor: | Phosducin-like protein 2, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
