2M51
 
 | |
2MHN
 
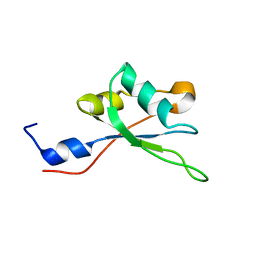 | |
2M52
 
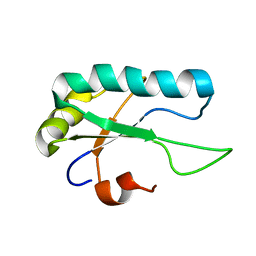 | |
2M7S
 
 | | NMR structure of RNA recognition motif 2 (RRM2) of Homo sapiens splicing factor, arginine/serine-rich 1 | | Descriptor: | Serine/arginine-rich splicing factor 1 | | Authors: | Dutta, S.K, Serrano, P, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG), Partnership for T-Cell Biology (TCELL) | | Deposit date: | 2013-04-29 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of RNA recognition motif 2 (RRM2) of Homo sapiens splicing factor, arginine/serine-rich 1
To be Published
|
|
2MCT
 
 | | NMR structure of the protein ZP_02042476.1 from Ruminococcus gnavus | | Descriptor: | Uncharacterized protein | | Authors: | Martin, B.T, Serrano, P, Geralt, M, Dutta, S, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2013-08-27 | | Release date: | 2013-11-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein ZP_02042476.1 from Ruminococcus gnavus.
To be Published
|
|
2MW1
 
 | |
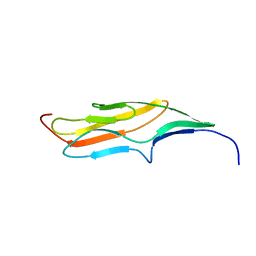
2MQC
 
 | |
2MQD
 
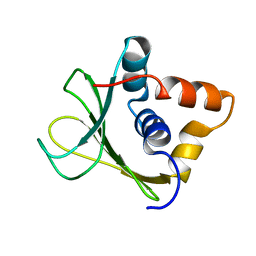 | |
2JZE
 
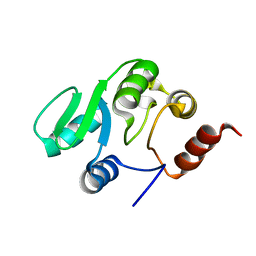 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3, single conformer closest to the mean coordinates of an ensemble of twenty energy minimized conformers | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2JZD
 
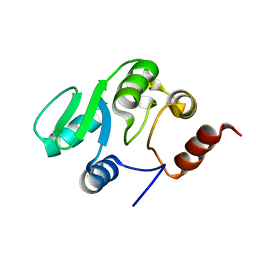 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2LGE
 
 | |
2ME0
 
 | |
2LXI
 
 | |
2M3D
 
 | |
2M26
 
 | |
2MCA
 
 | | NMR structure of the protein YP_002937094.1 from Eubacterium rectale | | Descriptor: | Uncharacterized protein | | Authors: | Proudfoot, A, Serrano, P, Geralt, M, Dutta, S, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein YP_002937094.1 from Eubacterium rectale
To be Published
|
|
2MVB
 
 | |
2MQB
 
 | |
2N6D
 
 | |
2MWM
 
 | |
2MXV
 
 | |
2MU1
 
 | | NMR structure of the core domain of NP_346487.1, a putative phosphoglycolate phosphatase from Streptococcus pneumoniae TIGR4 | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Jaudzems, K, Serrano, P, Pedrini, B, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-09-03 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | J-UNIO protocol used for NMR structure determination of the 206-residue protein NP_346487.1 from Streptococcus pneumoniae TIGR4.
J.Biomol.Nmr, 61, 2015
|
|
2MU2
 
 | | NMR structure of the cap domain of NP_346487.1, a putative phosphoglycolate phosphatase from Streptococcus pneumoniae TIGR4 | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Jaudzems, K, Serrano, P, Pedrini, B, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-09-03 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | J-UNIO protocol used for NMR structure determination of the 206-residue protein NP_346487.1 from Streptococcus pneumoniae TIGR4.
J.Biomol.Nmr, 61, 2015
|
|
2N8G
 
 | | NMR Structure of the homeodomain transcription factor Gbx1[E23R,R58E] from Homo sapiens | | Descriptor: | Homeobox protein GBX-1 | | Authors: | Proudfoot, A.K, Serrano, P, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG), Partnership for Stem Cell Biology (STEMCELL) | | Deposit date: | 2015-10-15 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the homeodomain transcription factor Gbx1[E23R,R58E] from Homo sapiens
To be Published
|
|
2N6E
 
 | |
