4JID
 
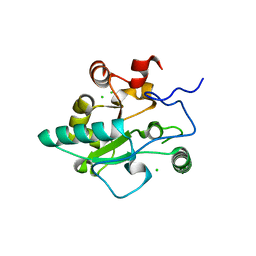 | | Crystal structure of BaLdcB / VanY-like L,D-carboxypeptidase Zinc(II)-free | | Descriptor: | CHLORIDE ION, D-alanyl-D-alanine carboxypeptidase family protein | | Authors: | Minasov, G, Wawrzak, Z, Onopriyenko, O, Skarina, T, Shatsman, S, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-05 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the LdcB LD-Carboxypeptidase Reveals the Molecular Basis of Peptidoglycan Recognition.
Structure, 22, 2014
|
|
3QFG
 
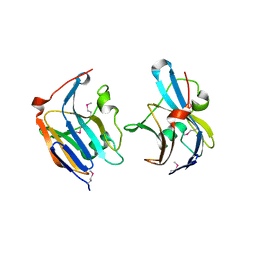 | | Structure of a putative lipoprotein from Staphylococcus aureus subsp. aureus NCTC 8325 | | Descriptor: | Uncharacterized protein | | Authors: | Filippova, E.V, Halavaty, A, Shuvalova, L, Minasov, G, Dubrovska, I, Winsor, J, Kiryukhina, O, Papazisi, L, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-21 | | Release date: | 2011-02-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structure of a putative lipoprotein from Staphylococcus aureus subsp. aureus NCTC 8325
To be Published
|
|
4JHX
 
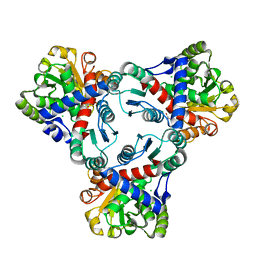 | | Crystal structure of anabolic ornithine carbamoyltransferase from Vibrio vulnificus in complex with carbamoylphosphate and arginine | | Descriptor: | ARGININE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shabalin, I.G, Bacal, P, Winsor, J, Grimshaw, S, Grabowski, M, Chordia, M.D, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-05 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures and kinetic properties of anabolic ornithine carbamoyltransferase from human pathogens Vibrio vulnificus and Bacillus anthracis
To be Published
|
|
4JLY
 
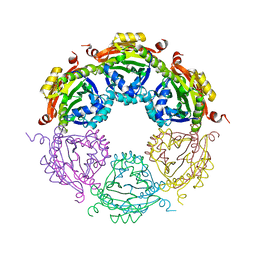 | | Dodecameric structure of spermidine N-acetyltransferase from Vibrio cholerae | | Descriptor: | SULFATE ION, Spermidine n1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.882 Å) | | Cite: | Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG.
J.Mol.Biol., 427, 2015
|
|
4GFP
 
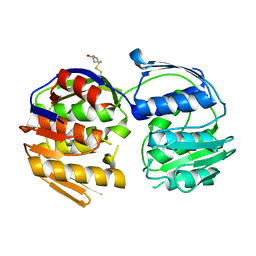 | | 2.7 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in a second conformational state | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, BETA-MERCAPTOETHANOL | | Authors: | Light, S.H, Minasov, G, Krishna, S.N, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-03 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 2.7 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in second conformational state
TO BE PUBLISHED
|
|
3RY3
 
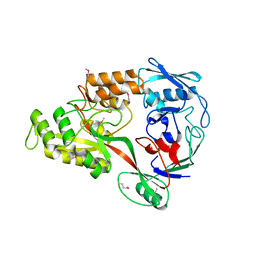 | | Putative solute-binding protein from Yersinia pestis. | | Descriptor: | ACETATE ION, Putative solute-binding protein | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-10 | | Release date: | 2011-05-18 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Putative solute-binding protein from Yersinia pestis.
To be Published
|
|
4HKJ
 
 | |
4FEZ
 
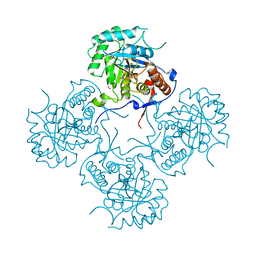 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant.
To be Published
|
|
4FFE
 
 | | The structure of cowpox virus CPXV018 (OMCP) | | Descriptor: | CPXV018 protein | | Authors: | Lazear, E, Peterson, L.W, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Cowpox Virus-Encoded NKG2D Ligand OMCP.
J.Virol., 87, 2013
|
|
4FG0
 
 | |
4FCA
 
 | | The crystal structure of a functionally unknown conserved protein from Bacillus anthracis str. Ames. | | Descriptor: | Conserved domain protein, IMIDAZOLE, NICKEL (II) ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-24 | | Release date: | 2012-06-06 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | The crystal structure of a functionally unknown conserved protein from Bacillus anthracis str. Ames.
To be Published
|
|
3S40
 
 | | The crystal structure of a diacylglycerol kinases from Bacillus anthracis str. Sterne | | Descriptor: | diacylglycerol kinase | | Authors: | Tan, K, Zhang, R, Xu, X, Cui, H, Peterson, S, Savchenko, A, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-18 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a diacylglycerol kinases from Bacillus anthracis str. Sterne
To be Published
|
|
3RHI
 
 | | DNA-binding protein HU from Bacillus anthracis | | Descriptor: | DNA-binding protein HU | | Authors: | Osipiuk, J, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | DNA-binding protein HU from Bacillus anthracis.
To be Published
|
|
3QDN
 
 | | Putative thioredoxin protein from Salmonella typhimurium | | Descriptor: | CALCIUM ION, Putative thioredoxin protein | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-18 | | Release date: | 2011-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Putative thioredoxin protein from Salmonella typhimurium.
To be Published
|
|
4ISX
 
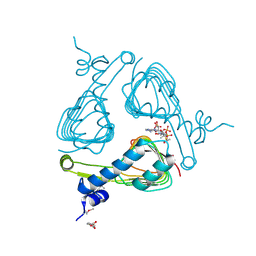 | | The crystal structure of maltose o-acetyltransferase from clostridium difficile 630 in complex with acetyl-coa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETYL COENZYME *A, Maltose O-acetyltransferase | | Authors: | Tan, K, Gu, G, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-17 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | The crystal structure of maltose o-acetyltransferase from clostridium difficile 630 in complex with acetyl-coa
To be Published
|
|
4JVO
 
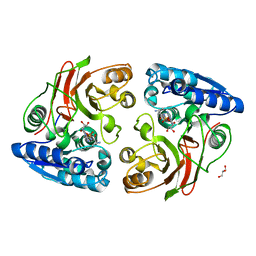 | | Crystal structure of the double mutant (S112A, H303A) of B.anthracis mycrocine immunity protein (MccF) with alanyl sulfamoyl adenylates | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, GLYCEROL, Microcin immunity protein MccF | | Authors: | Nocek, B, Tikhonov, A, Severinov, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-25 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the double mutant (S112A, H303A) of B.anthracis mycrocine immunity protein (MccF) with alanyl sulfamoyl adenylates
TO BE PUBLISHED
|
|
4JFR
 
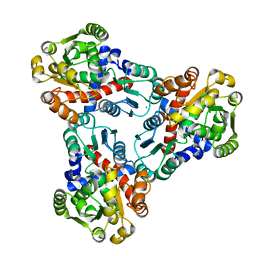 | | Crystal structure of anabolic ornithine carbamoyltransferase from Vibrio vulnificus in complex with carbamoyl phosphate | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Ornithine carbamoyltransferase, ... | | Authors: | Shabalin, I.G, Winsor, J, Grimshaw, S, Osinski, T, Bajor, J, Chordia, M.D, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-28 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structures and kinetic properties of anabolic ornithine carbamoyltransferase from human pathogens Vibrio vulnificus and Bacillus anthracis
to be published
|
|
4GB7
 
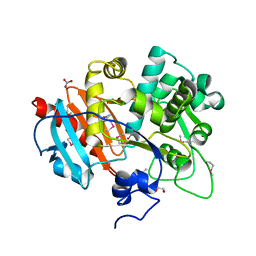 | | Putative 6-aminohexanoate-dimer hydrolase from Bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, 6-aminohexanoate-dimer hydrolase, NITRATE ION | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-07-26 | | Release date: | 2012-08-08 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Putative 6-aminohexanoate-dimer hydrolase from Bacillus anthracis.
To be Published
|
|
4FXS
 
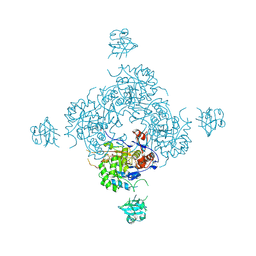 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae complexed with IMP and mycophenolic acid | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, MYCOPHENOLIC ACID, ... | | Authors: | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-07-03 | | Release date: | 2012-07-25 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae complexed with IMP and mycophenolic acid.
To be Published
|
|
4FO4
 
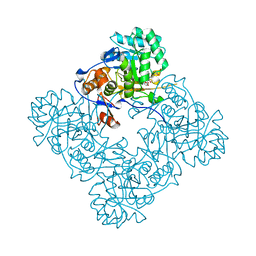 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, complexed with IMP and mycophenolic acid | | Descriptor: | INOSINIC ACID, Inosine 5'-monophosphate dehydrogenase, MYCOPHENOLIC ACID, ... | | Authors: | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, complexed with IMP and mycophenolic acid.
To be Published
|
|
4GL0
 
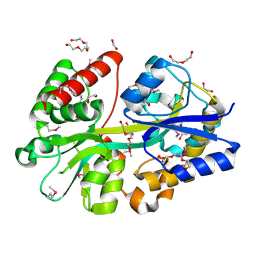 | | Putative spermidine/putrescine ABC transporter from Listeria monocytogenes | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Lmo0810 protein, ... | | Authors: | Osipiuk, J, Zhou, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-13 | | Release date: | 2012-08-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Putative spermidine/putrescine ABC transporter from Listeria monocytogenes
To be Published
|
|
3RDW
 
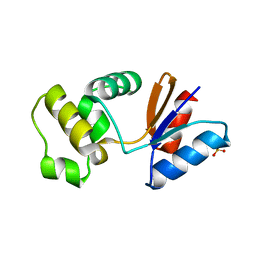 | | Putative arsenate reductase from Yersinia pestis | | Descriptor: | Putative arsenate reductase, SULFATE ION | | Authors: | Osipiuk, J, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-01 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Putative arsenate reductase from Yersinia pestis.
To be Published
|
|
4GVO
 
 | | Putative L-Cystine ABC transporter from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, HISTIDINE, Lmo2349 protein, ... | | Authors: | Osipiuk, J, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-30 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Putative L-Cystine ABC transporter from Listeria monocytogenes
To be Published
|
|
3R2E
 
 | | Dihydroneopterin aldolase/dihydroneopterin triphosphate 2'-epimerase from Yersinia pestis. | | Descriptor: | Dihydroneopterin aldolase | | Authors: | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-14 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dihydroneopterin aldolase/dihydroneopterin triphosphate 2'-epimerase from Yersinia pestis
To be Published
|
|
3Q88
 
 | | Glucose-6-phosphate isomerase from Francisella tularensis complexed with ribose 1,5-bisphosphate. | | Descriptor: | 1,5-di-O-phosphono-alpha-D-ribofuranose, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Osipiuk, J, Maltseva, N, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glucose-6-phosphate isomerase from Francisella tularensis.
To be Published
|
|
