8V4H
 
 | | X-ray structure of the NADP-dependent reductase from Campylobacter jejuni responsible for the synthesis of CDP-glucitol in the presence of CDP-glucitol | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Putative nucleotide sugar dehydratase, ... | | Authors: | Thoden, J.B, Schumann, M.E, Holden, H.M, Raushel, F.M. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biosynthesis of Cytidine Diphosphate-6-d-Glucitol for the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 63, 2024
|
|
4KCF
 
 | | X-ray Structure of a KijD3 in Complex with FMN and dTDP-3-amino-2,3,6-trideoxy-4-keto-3-methyl-D-glucose | | Descriptor: | FAD-dependent oxidoreductase, FLAVIN MONONUCLEOTIDE, [(2R,4S,6R)-4-azanyl-4,6-dimethyl-5,5-bis(oxidanyl)oxan-2-yl] [[(2R,3S,5R)-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] hydrogen phosphate | | Authors: | Holden, H.M, Thoden, J.B, Branch, M.C, Zimmer, A.L, Bruender, N.A. | | Deposit date: | 2013-04-24 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Active site architecture of a sugar N-oxygenase.
Biochemistry, 52, 2013
|
|
7US5
 
 | | X-ray crystal structure of GDP-D-glycero-D-manno-heptose 4,6-Dehydratase from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, GDP-D-GLYCERO-D-MANNO-HEPTOSE 4,6-DEHYDRATASE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-04-23 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reaction Mechanism and Three-Dimensional Structure of GDP-d-glycero-alpha-d-manno-heptose 4,6-Dehydratase from Campylobacter jejuni.
Biochemistry, 61, 2022
|
|
3ETJ
 
 | | Crystal structure E. coli Purk in complex with Mg, ADP, and Pi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, HYDROGENPHOSPHATE ION, ... | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2008-10-08 | | Release date: | 2008-10-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of the active site geometry of N(5)-Carboxyaminoimidazole ribonucleotide synthetase from Escherichia coli.
Biochemistry, 47, 2008
|
|
3ETH
 
 | | Crystal structure of E. coli Purk in complex with MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Phosphoribosylaminoimidazole carboxylase ATPase subunit | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2008-10-08 | | Release date: | 2008-10-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of the active site geometry of N(5)-Carboxyaminoimidazole ribonucleotide synthetase from Escherichia coli.
Biochemistry, 47, 2008
|
|
6CBM
 
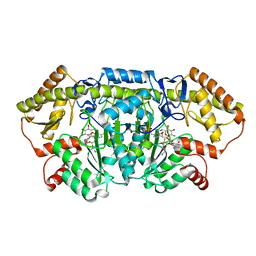 | | x-ray structure of NeoB from streptomyces fradiae in complex with PLP and neomycin (as the external aldimine) at pH 9 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-[(3-O-{2-amino-2,6-dideoxy-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-alpha-D-glucopyranosyl}-beta-D-ribofuranosyl)oxy]-3-hydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Dow, G.T, Holden, H.M. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
6CBK
 
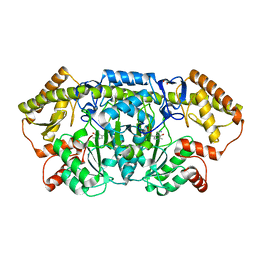 | | X-ray structure of NeoB from Streptomyces fradiae in complex with PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Neamine transaminase NeoN, ... | | Authors: | Thoden, J.B, Dow, G.T, Holden, H.M. | | Deposit date: | 2018-02-03 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
6CBL
 
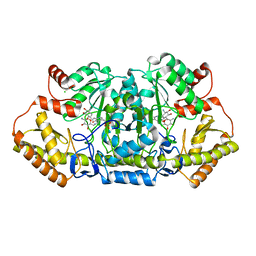 | | x-ray structure of NeoB from Streptomyces fradiae in complex with neamine as an external aldimine | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2-amino-2,6-dideoxy-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-alpha-D-glucopyranoside, CHLORIDE ION, Neamine transaminase NeoN | | Authors: | Thoden, J.B, Dow, G.T, Holden, H.M. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
6CBN
 
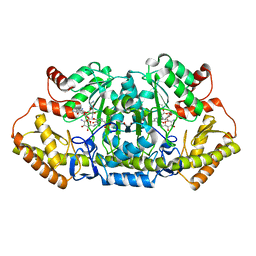 | | x-ray structure of NeoB from streptomyces fradiae in complex with PLP and neomycin (as the external aldimine) at pH 7.5 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-[(3-O-{2-amino-2,6-dideoxy-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-alpha-D-glucopyranosyl}-beta-D-ribofuranosyl)oxy]-3-hydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, 1,2-ETHANEDIOL, Neamine transaminase NeoN | | Authors: | Thoden, J.B, Dow, G.T, Holden, H.M. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
1BSL
 
 | |
2PA4
 
 | |
3OA0
 
 | | Crystal structure of the WlbA (WbpB) Dehydrogenase from Thermus thermophilus in complex with NAD and UDP-GlcNAcA | | Descriptor: | (2S,3S,4R,5R,6R)-5-acetamido-6-[[[(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-3,4-dihydroxy-oxane-2-carboxylic acid, CHLORIDE ION, Lipopolysaccharide biosynthesis protein wbpB, ... | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2010-08-04 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Studies of WlbA: A Dehydrogenase Involved in the Biosynthesis of 2,3-Diacetamido-2,3-dideoxy-d-mannuronic Acid .
Biochemistry, 49, 2010
|
|
3O9Z
 
 | | Crystal structure of the WlbA (WbpB) dehydrogenase from Thermus thermophilus in complex with NAD and alpha-ketoglutarate at 1.45 angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2010-08-04 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structural and Functional Studies of WlbA: A Dehydrogenase Involved in the Biosynthesis of 2,3-Diacetamido-2,3-dideoxy-d-mannuronic Acid .
Biochemistry, 49, 2010
|
|
3Q2K
 
 | | Crystal structure of the WlbA dehydrogenase from Bordetella pertussis in complex with NADH and UDP-GlcNAcA | | Descriptor: | (2S,3S,4R,5R,6R)-5-acetamido-6-[[[(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-3,4-dihydroxy-oxane-2-carboxylic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, oxidoreductase | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2010-12-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Biochemical and Structural Characterization of WlbA from Bordetella pertussis and Chromobacterium violaceum: Enzymes Required for the Biosynthesis of 2,3-Diacetamido-2,3-dideoxy-d-mannuronic Acid.
Biochemistry, 50, 2011
|
|
3Q2I
 
 | | Crystal structure of the WlbA dehydrognase from Chromobactrium violaceum in complex with NADH and UDP-GlcNAcA at 1.50 A resolution | | Descriptor: | (2S,3S,4R,5R,6R)-5-acetamido-6-[[[(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-3,4-dihydroxy-oxane-2-carboxylic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, ... | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2010-12-20 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and Structural Characterization of WlbA from Bordetella pertussis and Chromobacterium violaceum: Enzymes Required for the Biosynthesis of 2,3-Diacetamido-2,3-dideoxy-d-mannuronic Acid.
Biochemistry, 50, 2011
|
|
3OA2
 
 | |
5U21
 
 | | X-ray structure of the WlaRF aminotransferase from Campylobacter jejuni, K184A mutant in complex with TDP-Qui3N | | Descriptor: | (2R,3R,4S,5S,6R)-3,5-dihydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Holden, H.M, Dow, G.T, Gilbert, M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
5U20
 
 | | X-ray structure of the WlaRG aminotransferase from Campylobacter jejuni, internal PLP-aldimine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative aminotransferase, ... | | Authors: | Thoden, J.B, Holden, H.M, Dow, G.T, Gilbert, M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
5UIJ
 
 | | X-ray structure of The FdtF N-formyltransferase from Salmonella enteric O60 in complex with TDP | | Descriptor: | 1,2-ETHANEDIOL, Formyltransferase, SODIUM ION, ... | | Authors: | Thoden, J.B, Woodford, C.R, Holden, H.M. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular architecture of an N-formyltransferase from Salmonella enterica O60.
J. Struct. Biol., 200, 2017
|
|
5TIC
 
 | | X-ray structure of wild-type E. coli Acyl-CoA thioesterase I at pH 5 | | Descriptor: | Acyl-CoA thioesterase I, CHLORIDE ION | | Authors: | Thoden, J.B, Holden, H.M, Grisewood, M.J, Hernandez Lozada, N.J, Gifford, N.P, Mendez-Perez, D, Schoenberger, H.A, Allan, M.F, Pfleger, B.F, Marines, C.D. | | Deposit date: | 2016-10-02 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Computational Redesign of Acyl-ACP Thioesterase with Improved Selectivity toward Medium-Chain-Length Fatty Acids.
ACS Catal, 7, 2017
|
|
5TID
 
 | | X-ray structure of acyl-CoA thioesterase I, TesA, mutant M141L/Y145K/L146K at pH 5 in complex with octanoic acid | | Descriptor: | Acyl-CoA thioesterase I, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Thoden, J.B, Holden, H.M, Grisewood, M.J, Hernandez Lozada, N.J, Gifford, N.P, Mendez-Perez, D, Schoenberger, H.A, Allan, M.F, Pfleger, B.F, Marines, C.D. | | Deposit date: | 2016-10-02 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Computational Redesign of Acyl-ACP Thioesterase with Improved Selectivity toward Medium-Chain-Length Fatty Acids.
ACS Catal, 7, 2017
|
|
1KVU
 
 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-07 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic roles of tyrosine 149 and serine 124 in UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
1L7J
 
 | |
1L7K
 
 | |
1NS7
 
 | |
