4YVF
 
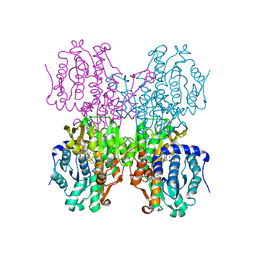 | | Structure of S-adenosyl-L-homocysteine hydrolase | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-{[5-chloro-2-(4-chlorophenoxy)phenyl](2-{[2-(methylamino)ethyl]amino}-2-oxoethyl)amino}-N-(1,3-dihydro-2H-isoindol-2-yl)-N-methylacetamide, Adenosylhomocysteinase | | Authors: | Akiko, K. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and structural analyses of S-adenosyl-L-homocysteine hydrolase inhibitors based on non-adenosine analogs.
Bioorg.Med.Chem., 23, 2015
|
|
2DW3
 
 | |
2QM6
 
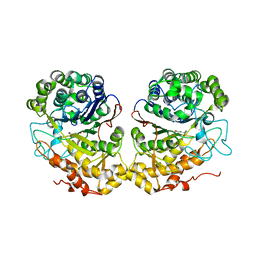 | |
5B5T
 
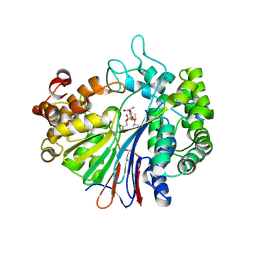 | | Crystal Structure of Escherichia coli Gamma-Glutamyltranspeptidase in Complex with peptidyl phosphonate inhibitor 1b | | Descriptor: | (2~{S})-2-azanyl-4-[(2~{R})-1-(2-hydroxy-2-oxoethylamino)-1-oxidanylidene-butan-2-yl]oxyphosphonoyl-butanoic acid, CALCIUM ION, Gamma-glutamyltranspeptidase large chain, ... | | Authors: | Wada, K, Fukuyama, K. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Phosphonate-based irreversible inhibitors of human gamma-glutamyl transpeptidase (GGT). GGsTop is a non-toxic and highly selective inhibitor with critical electrostatic interaction with an active-site residue Lys562 for enhanced inhibitory activity
Bioorg.Med.Chem., 24, 2016
|
|
5XDG
 
 | | Crystal structure of tertiary complex of TdsC from Paenibacillus sp. A11-2 with FMN and dibenzothiophene sulfoxide | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | Deposit date: | 2017-03-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
5XDE
 
 | | Crystal structure of tertiary complex of TdsC from Paenibacillus sp. A11-2 with FMN and dibenzothiophene | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | Deposit date: | 2017-03-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
5XB8
 
 | | Crystal structure of dibenzothiophene monooxygenase (TdsC) from Paenibacillus sp. A11-2 | | Descriptor: | SULFATE ION, Thermophilic dibenzothiophene desulfurization enzyme C | | Authors: | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
5XDD
 
 | | Crystal structure of tertiary complex of TdsC from Paenibacillus sp. A11-2 with FMN and Indole | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, INDOLE, ... | | Authors: | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | Deposit date: | 2017-03-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
5XDC
 
 | | Crystal structure of Indole-bound TdsC from Paenibacillus sp. A11-2 | | Descriptor: | GLYCEROL, INDOLE, SULFATE ION, ... | | Authors: | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | Deposit date: | 2017-03-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5785 Å) | | Cite: | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
5XDB
 
 | | Crystal structure of FMN-bound TdsC from Paenibacillus sp. A11-2 | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | Deposit date: | 2017-03-28 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
5YLV
 
 | | Crystal structure of the gastric proton pump complexed with SCH28080 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(2-methyl-8-phenylmethoxy-imidazo[1,2-a]pyridin-3-yl)ethanenitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abe, K, Irie, K, Nakanishi, H, Fujiyoshi, Y. | | Deposit date: | 2017-10-19 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79977775 Å) | | Cite: | Crystal structures of the gastric proton pump
Nature, 556, 2018
|
|
5YLU
 
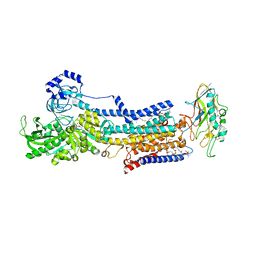 | | Crystal structure of the gastric proton pump complexed with vonoprazan | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[5-(2-fluorophenyl)-1-pyridin-3-ylsulfonyl-pyrrol-3-yl]-~{N}-methyl-methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abe, K, Irie, K, Nakanishi, H, Fujiyoshi, Y. | | Deposit date: | 2017-10-19 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79988956 Å) | | Cite: | Crystal structures of the gastric proton pump
Nature, 556, 2018
|
|
6AKG
 
 | |
6AKF
 
 | |
6AKE
 
 | |
2ZOY
 
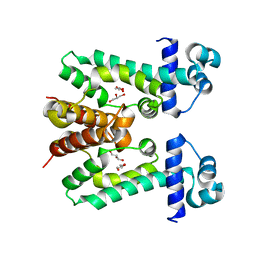 | | The multi-drug binding transcriptional repressor CgmR (CGL2612 protein) from C.glutamicum | | Descriptor: | GLYCEROL, Transcriptional regulator | | Authors: | Itou, H, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2008-06-20 | | Release date: | 2008-07-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The CGL2612 protein from Corynebacterium glutamicum is a drug resistance-related transcriptional repressor: structural and functional analysis of a newly identified transcription factor from genomic DNA analysis
J.Biol.Chem., 280, 2005
|
|
5YEC
 
 | | Crystal structure of Atg7CTD-Atg8-MgATP complex in form II | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Autophagy-related protein 8, MAGNESIUM ION, ... | | Authors: | Yamaguchi, M, Satoo, K, Noda, N.N. | | Deposit date: | 2017-09-16 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Atg7 Activates an Autophagy-Essential Ubiquitin-like Protein Atg8 through Multi-Step Recognition.
J. Mol. Biol., 430, 2018
|
|
2Z8I
 
 | |
2Z8J
 
 | |
2Z8K
 
 | |
7DTI
 
 | |
7DTH
 
 | | Solution structure of RPB6, common subunit of RNA polymerases I, II, and III | | Descriptor: | DNA-directed RNA polymerases I, II, and III subunit RPABC2 | | Authors: | Okuda, M, Nishimura, Y. | | Deposit date: | 2021-01-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three human RNA polymerases interact with TFIIH via a common RPB6 subunit.
Nucleic Acids Res., 50, 2022
|
|
3A75
 
 | |
5WZR
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - Gal-NHAc-DNJ complex | | Descriptor: | Alpha-N-acetylgalactosaminidase, CALCIUM ION, N-[(3S,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]acetamide, ... | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
3WHQ
 
 | |
