1P75
 
 | | Crystal structure of EHV4-TK complexed with TP5A | | Descriptor: | P1-(5'-ADENOSYL)P5-(5'-THYMIDYL)PENTAPHOSPHATE, SULFATE ION, Thymidine kinase | | Authors: | Gardberg, A, Shuvalova, L, Monnerjahn, C, Konrad, M, Lavie, A. | | Deposit date: | 2003-04-30 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural basis for the dual thymidine and thymidylate kinase activity of herpes thymidine kinases.
Structure, 11, 2003
|
|
1P7C
 
 | | Crystal Structure of HSV1-TK complexed with TP5A | | Descriptor: | P1-(5'-ADENOSYL)P5-(5'-THYMIDYL)PENTAPHOSPHATE, SULFATE ION, THYMIDINE, ... | | Authors: | Gardberg, A, Shuvalova, L, Monnerjahn, C, Konrad, M, Lavie, A. | | Deposit date: | 2003-05-01 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the dual thymidine and thymidylate kinase activity of herpes thymidine kinases.
Structure, 11, 2003
|
|
5EQV
 
 | | 1.45 Angstrom Crystal Structure of Bifunctional 2',3'-cyclic Nucleotide 2'-phosphodiesterase/3'-Nucleotidase Periplasmic Precursor Protein from Yersinia pestis with Phosphate bound to the Active site | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-MALATE, FE (III) ION, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-13 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 Angstrom Crystal Structure of Bifunctional 2',3'-cyclic Nucleotide 2'-phosphodiesterase/3'-Nucleotidase Periplasmic Precursor Protein from Yersinia pestis with Phosphate bound to the Active site.
To Be Published
|
|
1Q8I
 
 | | Crystal structure of ESCHERICHIA coli DNA Polymerase II | | Descriptor: | DNA polymerase II | | Authors: | Brunzelle, J.S, Muchmore, C.R.A, Mashhoon, N, Blair-Johnson, M, Shuvalova, L, Goodman, M.F, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-08-21 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Escherichia Coli DNA Polymerase II
To be Published
|
|
1P73
 
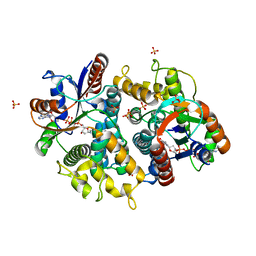 | | Crystal structure of EHV4-TK complexed with TP4A | | Descriptor: | P1-(5'-ADENOSYL)P4-(5'-(2'-DEOXY-THYMIDYL))TETRAPHOSPHATE, SULFATE ION, Thymidine kinase | | Authors: | Gardberg, A, Shuvalova, L, Monnerjahn, C, Konrad, M, Lavie, A. | | Deposit date: | 2003-04-30 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the dual thymidine and thymidylate kinase activity of herpes thymidine kinases.
Structure, 11, 2003
|
|
5DZS
 
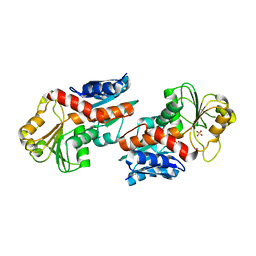 | | 1.5 Angstrom Crystal Structure of Shikimate Dehydrogenase 1 from Peptoclostridium difficile. | | Descriptor: | SULFATE ION, Shikimate dehydrogenase (NADP(+)) | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-26 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Shikimate Dehydrogenase 1 from Peptoclostridium difficile.
To Be Published
|
|
5HM8
 
 | | 2.85 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Adenosine and NAD. | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Halavaty, A, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-15 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 2.85 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Adenosine and NAD.
To Be Published
|
|
5HQH
 
 | | 1.32 Angstrom Crystal Structure of Ybbr like Domain of lmo2119 Protein from Listeria monocytogenes. | | Descriptor: | CHLORIDE ION, Lmo2119 protein, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-21 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | 1.32 Angstrom Crystal Structure of Ybbr like Domain of lmo2119 Protein from Listeria monocytogenes.
To Be Published
|
|
5HJ1
 
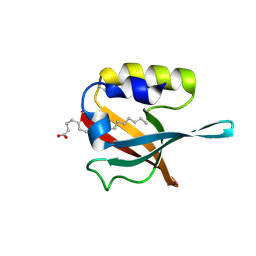 | | Crystal structure of PDZ domain of pullulanase C protein of type II secretion system from Klebsiella pneumoniae in complex with fatty acid | | Descriptor: | Pullulanase C protein, VACCENIC ACID | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of PDZ domain of pullulanase C protein of type II secretion system from Klebsiella pneumoniae in complex with fatty acid
To Be Published
|
|
4FCU
 
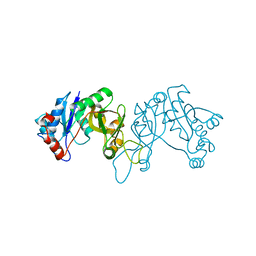 | | 1.9 Angstrom Crystal Structure of 3-deoxy-manno-octulosonate Cytidylyltransferase (kdsB) from Acinetobacter baumannii without His-Tag Bound to the Active Site | | Descriptor: | 3-deoxy-manno-octulosonate cytidylyltransferase | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-25 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Crystal Structure of 3-deoxy-manno-octulosonate Cytidylyltransferase (kdsB) from Acinetobacter baumannii without His-Tag Bound to the Active Site.
TO BE PUBLISHED
|
|
4FBD
 
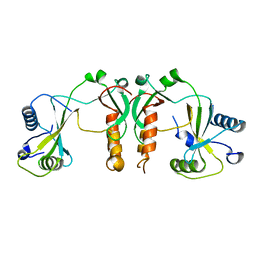 | | 2.35 Angstrom Crystal Structure of Conserved Hypothetical Protein from Toxoplasma gondii ME49. | | Descriptor: | Putative uncharacterized protein | | Authors: | Minasov, G, Ruan, J, Wawrzak, Z, Shuvalova, L, Ngo, H, Knoll, L, Milligan-Myhre, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom Crystal Structure of Conserved Hypothetical Protein from Toxoplasma gondii ME49.
TO BE PUBLISHED
|
|
1XA0
 
 | | Crystal Structure of MCSG Target APC35536 from Bacillus stearothermophilus | | Descriptor: | CHLORIDE ION, Putative NADPH Dependent oxidoreductases, SULFATE ION | | Authors: | Brunzelle, J.S, Sommerhalter, M, Minasov, G, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-24 | | Release date: | 2004-10-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of MCSG Target APC35536 from Bacillus stearothermophilus
To be Published
|
|
1YSJ
 
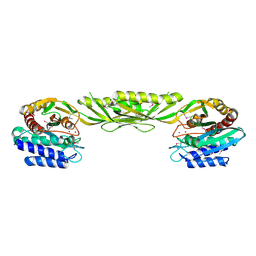 | | Crystal Structure of Bacillus Subtilis YXEP Protein (APC1829), a Dinuclear Metal Binding Peptidase from M20 Family | | Descriptor: | NICKEL (II) ION, protein yxeP | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-08 | | Release date: | 2005-03-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Bacillus Subtilis YXEP Protein, a Dinuclear Metal Binding Peptidase from M20 Family
To be Published
|
|
1ZSW
 
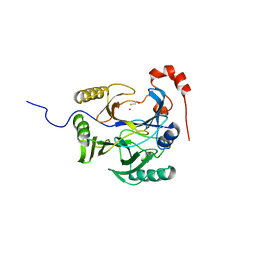 | | Crystal Structure of Bacillus cereus Metallo Protein from Glyoxalase family | | Descriptor: | BETA-MERCAPTOETHANOL, Glyoxalase family protein, ZINC ION | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-25 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Bacillus cereus Metallo Protein from Glyoxalase family
To be Published
|
|
6UOF
 
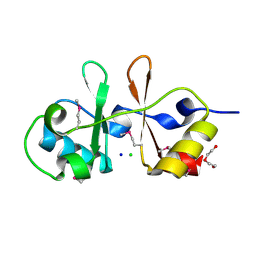 | | 1.2 Angstrom Resolution Crystal Structure of CBS Domains of Transcriptional Regulator from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-10-14 | | Release date: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 1.2 Angstrom Resolution Crystal Structure of CBS Domains of Transcriptional Regulator from Streptococcus pneumoniae
To Be Published
|
|
1ZKP
 
 | | 1.5A Resolution Crystal Structure of a Metallo Beta Lactamase Family Protein, the ELAC Homolgue of Bacillus anthracis, a Putative Ribonuclease | | Descriptor: | CHLORIDE ION, SODIUM ION, ZINC ION, ... | | Authors: | Brunzelle, J.S, Minasov, G, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | 1.5A Resolution Crystal Structure of a Metallo Beta Lactamase Family
Protein, the ELAC Homolgue of Bacillus anthracis, a Putative Ribonuclease
To be Published
|
|
6VBF
 
 | | 1.85 Angstrom Resolution Crystal Structure of N-terminal Domain of Two-component System Response Regulator from Acinetobacter baumannii | | Descriptor: | CALCIUM ION, CHLORIDE ION, Two-component regulatory system response regulator | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-18 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of N-terminal Domain of Two-component System Response Regulator from Acinetobacter baumannii
To Be Published
|
|
1Z05
 
 | | Crystal structure of the ROK family transcriptional regulator, homolog of E.coli MLC protein. | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, SULFATE ION, ... | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-01 | | Release date: | 2005-03-08 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the ROK family transcriptional regulator, homolog of E.coli MLC protein.
To be Published
|
|
6VJ6
 
 | | 2.55 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus cereus | | Descriptor: | 3,6,9,12,15-pentaoxaoctadecan-17-amine, GLYCEROL, Peptidylprolyl isomerase (PrsA) | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Wiersum, G, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-14 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | 2.55 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus cereus
To Be Published
|
|
1XPJ
 
 | | Crystal Structure of MCSG Target APC26283 from Vibrio cholerae | | Descriptor: | L(+)-TARTARIC ACID, MERCURY (II) ION, hypothetical protein | | Authors: | Brunzelle, J.S, Minasov, G, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-10-08 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of MCSG Target APC26283 from Vibrio cholerae
To be Published
|
|
6VBB
 
 | | 2.60 Angstrom Resolution Crystal Structure of Peptidase S41 from Acinetobacter baumannii | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Peptidase S41, ... | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-18 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.60 Angstrom Resolution Crystal Structure of Peptidase S41 from Acinetobacter baumannii
To Be Published
|
|
6W37
 
 | |
6UE2
 
 | | 1.85 Angstrom Resolution Crystal Structure of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-20 | | Release date: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of Class D beta-lactamase from Clostridium difficile 630.
To Be Published
|
|
6VJ4
 
 | | 1.70 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus anthracis | | Descriptor: | Peptidylprolyl isomerase PrsA | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Wiersum, G, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-14 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.70 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus anthracis
To Be Published
|
|
6VJ2
 
 | | 3.10 Angstrom Resolution Crystal Structure of Foldase Protein (PrsA) from Lactococcus lactis | | Descriptor: | Foldase | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-14 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | 3.10 Angstrom Resolution Crystal Structure of Foldase Protein (PrsA) from Lactococcus lactis
To Be Published
|
|
