8IAU
 
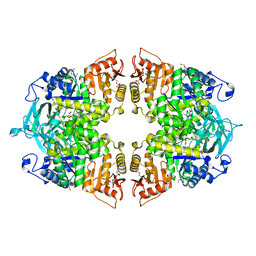 | | Crystal structure of Streptococcus pneumoniae pyruvate kinase in complex with oxalate and fructose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Nakashima, R, Taguchi, A. | | Deposit date: | 2023-02-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of Streptococcus pneumoniae pyruvate kinase involved in fosfomycin resistance.
J.Biol.Chem., 299, 2023
|
|
8IAV
 
 | |
8IAS
 
 | |
8IAW
 
 | |
8IAT
 
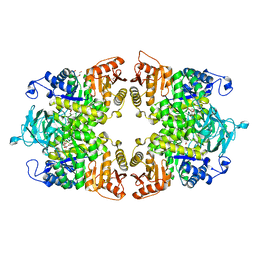 | |
8IAX
 
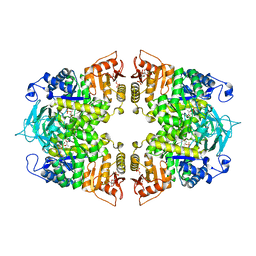 | | Crystal structure of Streptococcus pneumoniae pyruvate kinase in complex with phosphoenolpyruvate and fructose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Nakashima, R, Taguchi, A. | | Deposit date: | 2023-02-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and structural characterization of Streptococcus pneumoniae pyruvate kinase involved in fosfomycin resistance.
J.Biol.Chem., 299, 2023
|
|
7CAO
 
 | | Crystal structure of red chromoprotein from Olindias formosa | | Descriptor: | Chromoprotein | | Authors: | Nakashima, R, Zhai, L, Ike, Y, Matsudz, T, Nagai, T. | | Deposit date: | 2020-06-09 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-based analysis and evolution of a monomerized red-colored chromoprotein from the Olindias formosa jellyfish.
Protein Sci., 31, 2022
|
|
6IE8
 
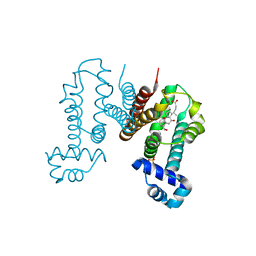 | | RamR in complex with cholic acid | | Descriptor: | CHOLIC ACID, Regulatory protein, SULFATE ION | | Authors: | Nakashima, R, Sakurai, K, Yamasaki, S, Nishino, K. | | Deposit date: | 2018-09-13 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the multidrug resistance regulator RamR complexed with bile acids.
Sci Rep, 9, 2019
|
|
6J1C
 
 | | Photoswitchable fluorescent protein Gamillus, N150C/T204V double mutant, off-state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein | | Authors: | Nakashima, R, Shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
6J1B
 
 | | Photoswitchable fluorescent protein Gamillus, N150C/T204V double mutant, on-state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
6JXF
 
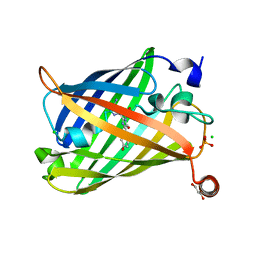 | | Photoswitchable fluorescent protein Gamillus, off-state (pH7.0) | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Sakurai, K, shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2019-04-23 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
6J1A
 
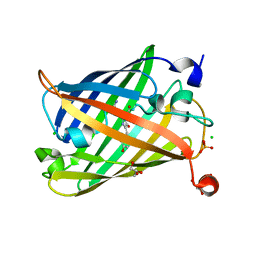 | | Photoswitchable fluorescent protein Gamillus, off-state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Sakurai, K, shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
6IE9
 
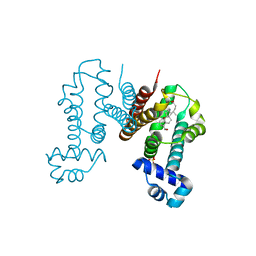 | | RamR in complex with chenodeoxycholic acid | | Descriptor: | CHENODEOXYCHOLIC ACID, Regulatory protein, SULFATE ION | | Authors: | Nakashima, R, Sakurai, K, Yamasaki, S, Nishino, K. | | Deposit date: | 2018-09-13 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the multidrug resistance regulator RamR complexed with bile acids.
Sci Rep, 9, 2019
|
|
8JMQ
 
 | | Crystal structure of hinokiresinol synthase | | Descriptor: | Hinokiresinol synthase alpha subunit, Hinokiresinol synthase beta subunit | | Authors: | Ding, Y, Ushimaru, R, Mori, T, Abe, I. | | Deposit date: | 2023-06-05 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insights into the C-C Bond-Forming Rearrangement Reaction Catalyzed by Heterodimeric Hinokiresinol Synthase.
J.Am.Chem.Soc., 145, 2023
|
|
8JMR
 
 | | Crystal structure of hinokiresinol synthase in complex with 1,7-bis(4-hydroxyphenyl)hepta-1,6-dien-3-one | | Descriptor: | 1,7-bis(4-hydroxyphenyl)hepta-1,6-dien-3-one, Hinokiresinol synthase alpha subunit, Hinokiresinol synthase beta subunit, ... | | Authors: | Ding, Y, Ushimaru, R, Mori, T, Abe, I. | | Deposit date: | 2023-06-05 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Insights into the C-C Bond-Forming Rearrangement Reaction Catalyzed by Heterodimeric Hinokiresinol Synthase.
J.Am.Chem.Soc., 145, 2023
|
|
1WD5
 
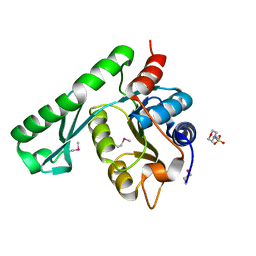 | | Crystal structure of TT1426 from Thermus thermophilus HB8 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, hypothetical protein TT1426 | | Authors: | Shibata, R, Kukimoto-Niino, M, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-11 | | Release date: | 2004-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a predicted phosphoribosyltransferase (TT1426) from Thermus thermophilus HB8 at 2.01 A resolution
Protein Sci., 14, 2005
|
|
6IIA
 
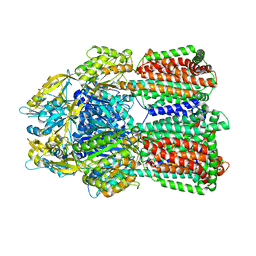 | | MexB in complex with LMNG | | Descriptor: | Lauryl Maltose Neopentyl Glycol, Multidrug resistance protein MexB | | Authors: | Nakashima, R, Sakurai, K, Nakao, K. | | Deposit date: | 2018-10-04 | | Release date: | 2019-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structures of multidrug efflux pump MexB bound with high-molecular-mass compounds.
Sci Rep, 9, 2019
|
|
2LAA
 
 | |
5Y01
 
 | | Acid-tolerant monomeric GFP, Gamillus, non-fluorescence (OFF) state | | Descriptor: | Green fluorescent protein, PHOSPHATE ION | | Authors: | Nakashima, R, Sakurai, K, Shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2017-07-14 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Acid-Tolerant Monomeric GFP from Olindias formosa.
Cell Chem Biol, 25, 2018
|
|
5Y00
 
 | | Acid-tolerant monomeric GFP, Gamillus, fluorescence (ON) state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Sakurai, K, Shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2017-07-14 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Acid-Tolerant Monomeric GFP from Olindias formosa.
Cell Chem Biol, 25, 2018
|
|
5XXN
 
 | | Crystal Structure of mutant (D286N) beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorose | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | Authors: | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahashi, Y, Sugimono, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
5XXM
 
 | | Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with gluconolactone | | Descriptor: | D-glucono-1,5-lactone, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | Authors: | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahash, Y, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
2CY1
 
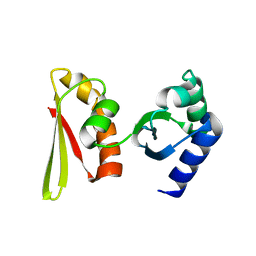 | | Crystal structure of APE1850 | | Descriptor: | NusA protein homolog | | Authors: | Shibata, R, Bessho, Y, Umehara, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization of the archaeal transcription termination factor NusA: a significant decrease in twinning under microgravity conditions
Acta Crystallogr.,Sect.F, 63, 2007
|
|
3W9I
 
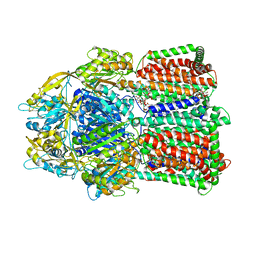 | | Structural basis for the inhibition of bacterial multidrug exporters | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Multidrug resistance protein MexB | | Authors: | Sakurai, K, Nakashima, R, Hayashi, K, Yamaguchi, A. | | Deposit date: | 2013-04-04 | | Release date: | 2013-07-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural basis for the inhibition of bacterial multidrug exporters
Nature, 500, 2013
|
|
3W9J
 
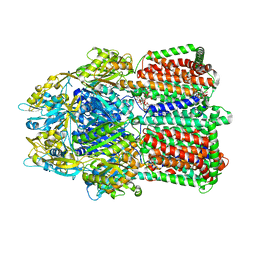 | | Structural basis for the inhibition of bacterial multidrug exporters | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Multidrug resistance protein MexB, [{2-[({[(3R)-1-{8-[(4-tert-butyl-1,3-thiazol-2-yl)carbamoyl]-4-oxo-3-[(E)-2-(1H-tetrazol-5-yl)ethenyl]-4H-pyrido[1,2-a]pyrimidin-2-yl}piperidin-3-yl]oxy}carbonyl)amino]ethyl}(dimethyl)ammonio]acetate | | Authors: | Sakurai, K, Nakashima, R, Hayashi, K, Yamaguchi, A. | | Deposit date: | 2013-04-04 | | Release date: | 2013-07-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for the inhibition of bacterial multidrug exporters
Nature, 500, 2013
|
|
