3VVO
 
 | |
3VVN
 
 | |
3WBN
 
 | | Crystal structure of MATE in complex with MaL6 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MaL6, Putative uncharacterized protein | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-20 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3W4T
 
 | | Crystal structure of MATE P26A mutant | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative uncharacterized protein | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-01-16 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
7VZP
 
 | | FAD-dpendent Glucose Dehydrogenase from Aspergillus oryzae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GMC oxidoreductase, PENTAETHYLENE GLYCOL | | Authors: | Nakajima, Y. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational change of catalytic residue in reduced enzyme of FAD-dependent Glucose Dehydrogenase at pH6.5
To Be Published
|
|
7VZS
 
 | | FAD-dpendent Glucose Dehydrogenase complexed with an inhibitor at pH7.56 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-glucal, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nakajima, Y. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational change of catalytic residue in reduced enzyme of FAD-dependent Glucose Dehydrogenase at pH6.5
To Be Published
|
|
3VYG
 
 | | Crystal structure of Thiocyanate hydrolase mutant R136W | | Descriptor: | COBALT (III) ION, L(+)-TARTARIC ACID, Thiocyanate hydrolase subunit alpha, ... | | Authors: | Yamanaka, Y, Sato, M, Arakawa, T, Namima, S, Hori, S, Ohtaki, A, Noguchi, K, Katayama, Y, Yohda, M, Odaka, M. | | Deposit date: | 2012-09-25 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Effects of argnine residue around the substrate pocket on the substrate specificity of thiocyanate hydrolase
To be published
|
|
3VXD
 
 | | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N from Streptcoccus agalactiae | | Descriptor: | Putative uncharacterized protein gbs1889, SULFATE ION | | Authors: | Nakamichi, Y, Maruyama, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2012-09-11 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N from Streptcoccus agalactiae
To be Published
|
|
5X70
 
 | |
5X6X
 
 | |
5X6Z
 
 | |
5X6Y
 
 | |
5X71
 
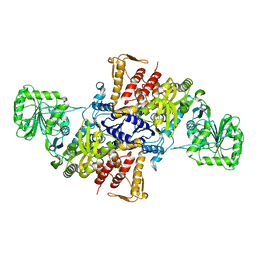 | |
5ZM4
 
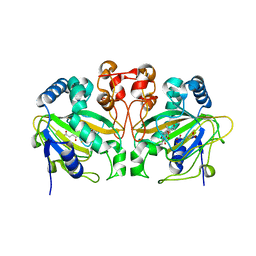 | | Fe(II)/(alpha)ketoglutarate-dependent dioxygenase AndA with preandiloid C | | Descriptor: | (6aS,8aR,12aS,12bR,13aR)-5,6a,9,9,12a,13a-hexamethyl-7,8,8a,9,12a,12b,13,13a-octahydro-3H-benzo[a]furo[3,4-j]xanthene-3,4,10(1H,6aH)-trione, 2-OXOGLUTARIC ACID, Dioxygenase andA, ... | | Authors: | Nakashima, Y, Senda, T. | | Deposit date: | 2018-04-01 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Computational Bases for Dramatic Skeletal Rearrangement in Anditomin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
5ZM2
 
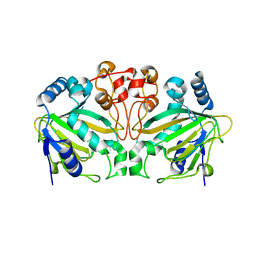 | |
7DQK
 
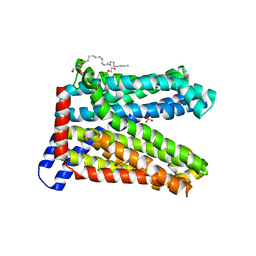 | | A nicotine MATE transporter, Nicotiana tabacum MATE2 (NtMATE2) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanaka, Y, Tsukazaki, T, Sasaki, A, Iwaki, S. | | Deposit date: | 2020-12-24 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of a nicotine MATE transporter provide insight into its mechanism of substrate transport.
Febs Lett., 595, 2021
|
|
2YXX
 
 | |
3WUX
 
 | | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N/K370S from Streptococcus agalactiae | | Descriptor: | 1,2-ETHANEDIOL, Unsaturated chondroitin disaccharide hydrolase | | Authors: | Nakamichi, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-05-08 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N/K370S from Streptococcus agalactiae
to be published
|
|
3WIW
 
 | | Crystal structure of unsaturated glucuronyl hydrolase specific for heparin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Glycosyl hydrolase family 88 | | Authors: | Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2013-09-26 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a bacterial unsaturated glucuronyl hydrolase with specificity for heparin.
J.Biol.Chem., 289, 2014
|
|
3X28
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K | | Descriptor: | CHLORIDE ION, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2014-12-12 | | Release date: | 2015-12-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Reaction intermediate of nitrile hydratase determined by time-resolved crystallography reveals the cysteine-sulfenic acid ligand to be a catalytic nucleophile
To be Published
|
|
2YYR
 
 | |
3ANI
 
 | | Crystal structure of unsaturated glucuronyl hydrolase mutant D175N from Streptcoccus agalactiae | | Descriptor: | Putative uncharacterized protein gbs1889 | | Authors: | Nakamichi, Y, Maruyama, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2010-09-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural determinants in streptococcal unsaturated glucuronyl hydrolase for recognition of glycosaminoglycan sulfate groups
J.Biol.Chem., 286, 2011
|
|
3ANK
 
 | | Crystal structure of unsaturated glucuronyl hydrolase mutant D175N from Streptcoccus agalactiae complexed with dGlcA-GalNAc6S | | Descriptor: | 1,2-ETHANEDIOL, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-6-O-sulfo-beta-D-galactopyranose, Putative uncharacterized protein gbs1889 | | Authors: | Nakamichi, Y, Maruyama, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2010-09-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural determinants in streptococcal unsaturated glucuronyl hydrolase for recognition of glycosaminoglycan sulfate groups
J.Biol.Chem., 286, 2011
|
|
3ANJ
 
 | | Crystal structure of unsaturated glucuronyl hydrolase from Streptcoccus agalactiae | | Descriptor: | Putative uncharacterized protein gbs1889 | | Authors: | Nakamichi, Y, Maruyama, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2010-09-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural determinants in streptococcal unsaturated glucuronyl hydrolase for recognition of glycosaminoglycan sulfate groups
J.Biol.Chem., 286, 2011
|
|
3A8G
 
 | | Crystal structure of Nitrile Hydratase mutant S113A complexed with Trimethylacetonitrile | | Descriptor: | 2,2-dimethylpropanenitrile, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Ohtaki, A, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2009-10-06 | | Release date: | 2010-04-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Kinetic and structural studies on roles of the serine ligand and a strictly conserved tyrosine residue in nitrile hydratase
J.Biol.Inorg.Chem., 15, 2010
|
|
