3KP1
 
 | | Crystal structure of ornithine 4,5 aminomutase (Resting State) | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, D-ornithine aminomutase E component, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KOW
 
 | | Crystal Structure of ornithine 4,5 aminomutase backsoaked complex | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, D-ornithine aminomutase E component, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
4YEM
 
 | | Carboplatin binding to HEWL in NaBr crystallisation conditions studied at an X-ray wavelength of 0.9163A - new refinement | | Descriptor: | ACETATE ION, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7A9I
 
 | |
7A9J
 
 | |
6S7E
 
 | | Plant Cysteine Oxidase PCO4 from Arabidopsis thaliana (using PEG 3350 and NaF as precipitants) | | Descriptor: | FE (III) ION, Plant cysteine oxidase 4 | | Authors: | White, M.D, Levy, C.W, Flashman, E, McDonough, M.A. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of Arabidopsis thaliana oxygen-sensing plant cysteine oxidases 4 and 5 enable targeted manipulation of their activity.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7ZP7
 
 | | Crystal structure of evolved photoenzyme EnT1.3 (truncated) with bound product | | Descriptor: | (1~{R},10~{R},12~{S})-15-oxa-8-azatetracyclo[8.5.0.0^{1,12}.0^{2,7}]pentadeca-2(7),3,5-trien-9-one, 1,2-ETHANEDIOL, EnT1.3 C | | Authors: | Hardy, F.J, Levy, C. | | Deposit date: | 2022-04-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A designed photoenzyme for enantioselective [2+2] cycloadditions.
Nature, 611, 2022
|
|
5M0O
 
 | | Crystal structure of cytochrome P450 OleT H85Q in complex with arachidonic acid | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Tee, K.L, Munro, A, Matthews, S, Leys, D, Levy, C. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic Determinants of Alkene Production by the Cytochrome P450 Peroxygenase OleTJE.
J. Biol. Chem., 292, 2017
|
|
5M0P
 
 | | Crystal structure of cytochrome P450 OleT F79A in complex with arachidonic acid | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, Terminal olefin-forming fatty acid decarboxylase, ... | | Authors: | Tee, K.L, Munro, A, Matthews, S, Leys, D, Levy, C. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Catalytic Determinants of Alkene Production by the Cytochrome P450 Peroxygenase OleTJE.
J. Biol. Chem., 292, 2017
|
|
5M0N
 
 | | Crystal structure of cytochrome P450 OleT in complex with formate | | Descriptor: | FORMIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Tee, K.L, Munro, A, Matthews, S, Leys, D, Levy, C. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Catalytic Determinants of Alkene Production by the Cytochrome P450 Peroxygenase OleTJE.
J. Biol. Chem., 292, 2017
|
|
7ZP6
 
 | |
7ZP5
 
 | |
4RAS
 
 | | Reductive dehalogenase structure suggests a mechanism for B12-dependent dehalogenation | | Descriptor: | CHLORIDE ION, COBALAMIN, IRON/SULFUR CLUSTER, ... | | Authors: | Quezada, C.P, Payne, K.A.P, Leys, D. | | Deposit date: | 2014-09-11 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reductive dehalogenase structure suggests a mechanism for B12-dependent dehalogenation.
Nature, 517, 2015
|
|
6RQ5
 
 | | CYP121 in complex with 3,5-dimethyl dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(3,5-dimethyl-4-oxidanyl-phenyl)methyl]-6-[(4-hydroxyphenyl)methyl]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mycocyclosin synthase, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6RQE
 
 | | CYP121 in complex with 3-acetylene dicyclotyrosine | | Descriptor: | 3-acetylene dicyclotyrosine, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure-Activity Relationships ofcyclo(l-Tyrosyl-l-tyrosine) Derivatives Binding toMycobacterium tuberculosisCYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6RQ8
 
 | | CYP121 in complex with 3-iodo dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(4-hydroxyphenyl)methyl]-6-[(3-iodanyl-4-oxidanyl-phenyl)methyl]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mycocyclosin synthase, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6RQ0
 
 | | CYP121 in complex with 3-methyl dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(4-hydroxyphenyl)methyl]-6-[(3-methyl-4-oxidanyl-phenyl)methyl]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mycocyclosin synthase, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6RQD
 
 | | CYP121 in complex with 3-chloro dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(3-chloranyl-4-oxidanyl-phenyl)methyl]-6-[(4-hydroxyphenyl)methyl]piperazine-2,5-dione, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6RQ9
 
 | | CYP121 in complex with O-methyl dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(4-hydroxyphenyl)methyl]-6-[(4-methoxyphenyl)methyl]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mycocyclosin synthase, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6RQ6
 
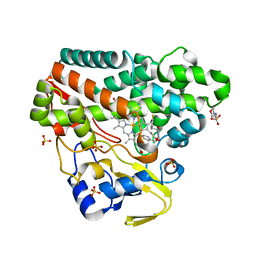 | | CYP121 in complex with 3-fluoro dicyclotyrosine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-fluoro dicyclotyrosine, Mycocyclosin synthase, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6RQ1
 
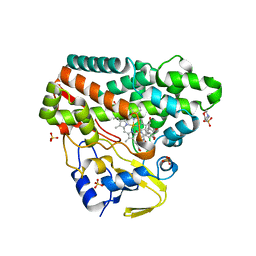 | | CYP121 in complex with 2-methyl dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(4-hydroxyphenyl)methyl]-6-[(2-methyl-4-oxidanyl-phenyl)methyl]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mycocyclosin synthase, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6RQB
 
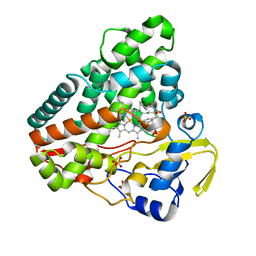 | | CYP121 in complex with 3-bromo dicyclotyrosine | | Descriptor: | 3-bromo dicyclotyrosine, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.459 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6RQ3
 
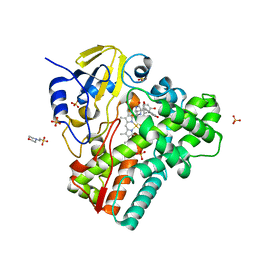 | | CYP121 in complex with 2,6-dimethyl dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(2,6-dimethyl-4-oxidanyl-phenyl)methyl]-6-[(4-hydroxyphenyl)methyl]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mycocyclosin synthase, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
4LT3
 
 | | HEWL co-crystallized with Carboplatin in non-NaCl conditions: crystal 2 processed using the XDS software package | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Diederichs, K, Kroon-Batenburg, L.M.J, Schreurs, A.M.M, Helliwell, J.R. | | Deposit date: | 2013-07-23 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carboplatin binding to histidine.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4LT0
 
 | | HEWL co-crystallized with Carboplatin in non-NaCl conditions: crystal 1 processed using the EVAL software package | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Diederichs, K, Kroon-Batenburg, L.M.J, Schreurs, A.M.M, Helliwell, J.R. | | Deposit date: | 2013-07-23 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Carboplatin binding to histidine.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
