1CAV
 
 | |
7V6H
 
 | | Crystal Structure of the SpnL | | Descriptor: | Cyclopropane fatty-acyl-phospholipid synthase-like methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wu, H.-H, Ko, T.-P, Liu, H.-W, Tsai, M.-D. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.054 Å) | | Cite: | Evidence for an Enzyme-Catalyzed Rauhut-Currier Reaction during the Biosynthesis of Spinosyn A.
J.Am.Chem.Soc., 143, 2021
|
|
2D45
 
 | | Crystal structure of the MecI-mecA repressor-operator complex | | Descriptor: | 5'-D(P*TP*AP*CP*TP*AP*CP*AP*TP*AP*TP*GP*TP*AP*GP*TP*A)-3', Methicillin resistance regulatory protein mecI | | Authors: | Safo, M.K, Ko, T.-P, Musayev, F.N, Zhao, Q, Wang, A.H.-J, Archer, G.L. | | Deposit date: | 2005-10-09 | | Release date: | 2005-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the MecI repressor from Staphylococcus aureus in complex with the cognate DNA operator of mec.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
7YQN
 
 | | Crystal structure of Ecoli malate synthase G | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wu, K.-P, Lu, Y.-C, Ko, T.-P. | | Deposit date: | 2022-08-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cryo-EM reveals the structure and dynamics of a 723-residue malate synthase G.
J.Struct.Biol., 215, 2023
|
|
8J0A
 
 | | Robust design of effective allosteric activator UbV R4 for Rsp5 E3 ligase using the machine-learning tool ProteinMPNN | | Descriptor: | SULFATE ION, Ubiquitin variant R4 | | Authors: | Lin, Y.-F, Hsieh, Y.-J, Kao, H.-W, Ko, T.-P, Wu, K.-P. | | Deposit date: | 2023-04-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Robust Design of Effective Allosteric Activators for Rsp5 E3 Ligase Using the Machine Learning Tool ProteinMPNN.
Acs Synth Biol, 12, 2023
|
|
3RNZ
 
 | |
3RO1
 
 | |
3RO0
 
 | |
1SD7
 
 | | Crystal Structure of a SeMet derivative of MecI at 2.65 A | | Descriptor: | Methicillin resistance regulatory protein mecI | | Authors: | Safo, M.K, Zhao, Q, Musayev, F.N, Robinson, H, Scarsdale, N, Archer, G.L. | | Deposit date: | 2004-02-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of the BlaI repressor from Staphylococcus aureus and its complex with DNA: insights into transcriptional regulation of the bla and mec operons
J.Bacteriol., 187, 2005
|
|
3PJG
 
 | | Crystal structure of UDP-glucose dehydrogenase from Klebsiella pneumoniae complexed with product UDP-glucuronic acid | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, UDP-glucose 6-dehydrogenase, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Chen, Y.-Y, Ko, T.-P, Lin, C.-H, Chen, W.-H, Wang, A.H.-J. | | Deposit date: | 2010-11-10 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
3PID
 
 | | The apo-form UDP-glucose 6-dehydrogenase with a C-terminal six-histidine tag | | Descriptor: | UDP-glucose 6-dehydrogenase | | Authors: | Chen, Y.-Y, Ko, T.-P, Lin, C.-H, Chen, W.-H, Wang, A.H.-J. | | Deposit date: | 2010-11-06 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
3PLN
 
 | | Crystal structure of Klebsiella pneumoniae UDP-glucose 6-dehydrogenase complexed with UDP-glucose | | Descriptor: | UDP-glucose 6-dehydrogenase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Chen, Y.-Y, Ko, T.-P, Lin, C.-H, Chen, W.-H, Wang, A.H.-J. | | Deposit date: | 2010-11-15 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
3PLR
 
 | | Crystal structure of Klebsiella pneumoniae UDP-glucose 6-dehydrogenase complexed with NADH and UDP-glucose | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, UDP-glucose 6-dehydrogenase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Chen, Y.-Y, Ko, T.-P, Lin, C.-H, Chen, W.-H, Wang, A.H.-J. | | Deposit date: | 2010-11-15 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
1SD4
 
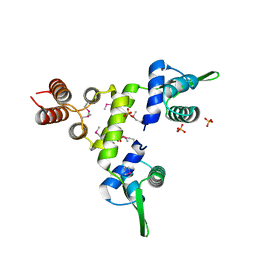 | | Crystal Structure of a SeMet derivative of BlaI at 2.0 A | | Descriptor: | PENICILLINASE REPRESSOR, SULFATE ION | | Authors: | Safo, M.K, Zhao, Q, Musayev, F.N, Robinson, H, Scarsdale, N, Archer, G.L. | | Deposit date: | 2004-02-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the BlaI repressor from Staphylococcus aureus and its complex with DNA: insights into transcriptional regulation of the bla and mec operons
J.Bacteriol., 187, 2005
|
|
1SD6
 
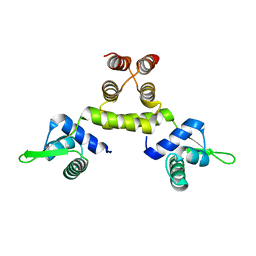 | | Crystal Structure of Native MecI at 2.65 A | | Descriptor: | Methicillin resistance regulatory protein mecI | | Authors: | Safo, M.K, Zhao, Q, Musayev, F.N, Robinson, H, Scarsdale, N, Archer, G.L. | | Deposit date: | 2004-02-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of the BlaI repressor from Staphylococcus aureus and its complex with DNA: insights into transcriptional regulation of the bla and mec operons
J.Bacteriol., 187, 2005
|
|
7D4U
 
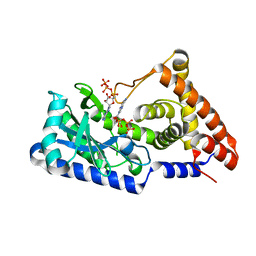 | | ATP complex with double mutant cyclic trinucleotide synthase CdnD | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclic AMP-AMP-GMP synthase | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7D4S
 
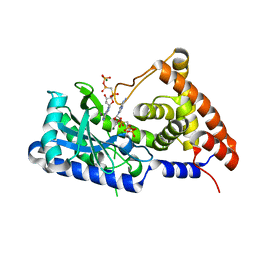 | | apo-form cyclic trinucleotide synthase CdnD | | Descriptor: | Cyclic AMP-AMP-GMP synthase, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7D48
 
 | | apo-form cyclic trinucleotide synthase CdnD | | Descriptor: | Cyclic AMP-AMP-GMP synthase, SODIUM ION | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-23 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7D4J
 
 | | ddATP complex of cyclic trinucleotide synthase CdnD | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, Cyclic AMP-AMP-GMP synthase, MAGNESIUM ION | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7D4O
 
 | | cyclic trinucleotide synthase CdnD in complex with ATP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cyclic AMP-AMP-GMP synthase, ... | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
1V4L
 
 | |
4NI0
 
 | | Quaternary R3 CO-liganded hemoglobin structure in complex with a thiol containing compound | | Descriptor: | 5-[(2S)-2,3-dihydro-1,4-benzodioxin-2-yl]-2,4-dihydro-3H-1,2,4-triazole-3-thione, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Safo, M.K, Meadows, J, Ko, T.-P, Nakagawa, A, Zapol, W. | | Deposit date: | 2013-11-05 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of a Small Molecule that Increases Hemoglobin Oxygen Affinity and Reduces SS Erythrocyte Sickling.
Acs Chem.Biol., 9, 2014
|
|
4NI1
 
 | | Quaternary R CO-liganded hemoglobin structure in complex with a thiol containing compound | | Descriptor: | 5-[(2R)-2,3-dihydro-1,4-benzodioxin-2-yl]-2,4-dihydro-3H-1,2,4-triazole-3-thione, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Safo, M.K, Meadows, J, Ko, T.-P, Nakagawa, A, Zapol, W. | | Deposit date: | 2013-11-05 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a Small Molecule that Increases Hemoglobin Oxygen Affinity and Reduces SS Erythrocyte Sickling.
Acs Chem.Biol., 9, 2014
|
|
1X06
 
 | | Crystal structure of undecaprenyl pyrophosphate synthase in complex with Mg, IPP and Fspp | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, ... | | Authors: | Guo, R.-T, Ko, T.-P, Chen, A.P.-C, Kuo, C.-J, Wang, A.H.-J, Liang, P.-H. | | Deposit date: | 2005-03-15 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of undecaprenyl pyrophosphate synthase in complex with magnesium, isopentenyl pyrophosphate, and farnesyl thiopyrophosphate: roles of the metal ion and conserved residues in catalysis.
J.Biol.Chem., 280, 2005
|
|
1X07
 
 | | Crystal structure of undecaprenyl pyrophosphate synthase in complex with Mg and IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Guo, R.-T, Ko, T.-P, Chen, A.P.-C, Kuo, C.-J, Wang, A.H.-J, Liang, P.-H. | | Deposit date: | 2005-03-15 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of undecaprenyl pyrophosphate synthase in complex with magnesium, isopentenyl pyrophosphate, and farnesyl thiopyrophosphate: roles of the metal ion and conserved residues in catalysis.
J.Biol.Chem., 280, 2005
|
|
