8KD9
 
 | | Cryo-EM structure of Aquifex aeolicus minimal protein-only RNase P (HARP) in complex with pre-tRNAs | | Descriptor: | Aquifex aeolicus pre-tRNAVal, RNA-free ribonuclease P | | Authors: | Teramoto, T, Koyasu, T, Mayanagi, K, Yokogawa, T, Adachi, N, Nakamura, T, Senda, T, Kakuta, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM structure of Aquifex aeolicus minimal protein-only RNase P (HARP) in complex with pre-tRNAs
To Be Published
|
|
2D6P
 
 | | Crystal structure of mouse galectin-9 N-terminal CRD in complex with T-antigen | | Descriptor: | beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, lectin, galactose binding, ... | | Authors: | Nagae, M, Nishi, N, Nakamura, T, Murata, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Galectin-9 N-terminal Carbohydrate Recognition Domain from Mus musculus Reveals the Basic Mechanism of Carbohydrate Recognition
J.Biol.Chem., 281, 2006
|
|
2D6N
 
 | | Crystal structure of mouse galectin-9 N-terminal CRD in complex with N-acetyllactosamine | | Descriptor: | beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, lectin, galactose binding, ... | | Authors: | Nagae, M, Nishi, N, Nakamura, T, Murata, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Galectin-9 N-terminal Carbohydrate Recognition Domain from Mus musculus Reveals the Basic Mechanism of Carbohydrate Recognition
J.Biol.Chem., 281, 2006
|
|
2D6O
 
 | | Crystal structure of mouse galectin-9 N-terminal CRD in complex with N-acetyllactosamine dimer | | Descriptor: | GLYCEROL, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, lectin, ... | | Authors: | Nagae, M, Nishi, N, Nakamura, T, Murata, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-09-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of the Galectin-9 N-terminal Carbohydrate Recognition Domain from Mus musculus Reveals the Basic Mechanism of Carbohydrate Recognition
J.Biol.Chem., 281, 2006
|
|
5XQY
 
 | |
5XP1
 
 | |
5LAO
 
 | | S-nitrosylated 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5LAM
 
 | | Refined 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
6L0R
 
 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with O-Acetylserine | | Descriptor: | (2S)-3-acetyloxy-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | Authors: | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
6L0S
 
 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with L-Cysteine | | Descriptor: | (2R)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-sulfanyl-propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | Authors: | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
6M5Y
 
 | |
6L0Q
 
 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with O-Phosphoserine | | Descriptor: | (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-phosphonooxy-propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | Authors: | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
6L0P
 
 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with O-Phosphoserine | | Descriptor: | (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-phosphonooxy-propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | Authors: | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
2DFV
 
 | | Hyperthermophilic threonine dehydrogenase from Pyrococcus horikoshii | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Probable L-threonine 3-dehydrogenase, SULFATE ION, ... | | Authors: | Ishikawa, K, Higashi, N, Nakamura, T, Matsuura, T, Nakagawa, A. | | Deposit date: | 2006-03-03 | | Release date: | 2007-01-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The first crystal structure of L-threonine dehydrogenase.
J.Mol.Biol., 366, 2007
|
|
6KRP
 
 | |
6KRQ
 
 | |
6KRK
 
 | |
3APV
 
 | | Crystal structure of the A variant of human alpha1-acid glycoprotein and amitriptyline complex | | Descriptor: | ACETIC ACID, Alpha-1-acid glycoprotein 2, Amitriptyline | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
6KRR
 
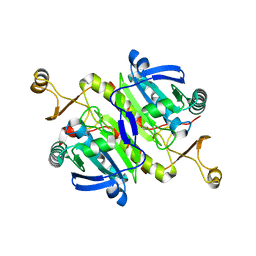 | |
3APX
 
 | | Crystal structure of the A variant of human alpha1-acid glycoprotein and chlorpromazine complex | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, ACETIC ACID, Alpha-1-acid glycoprotein 2 | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
3APW
 
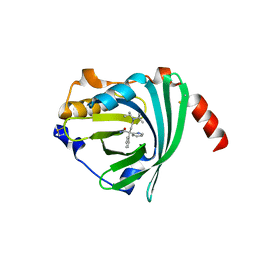 | | Crystal structure of the A variant of human alpha1-acid glycoprotein and disopyramide complex | | Descriptor: | Alpha-1-acid glycoprotein 2, Disopyramide | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
6KRM
 
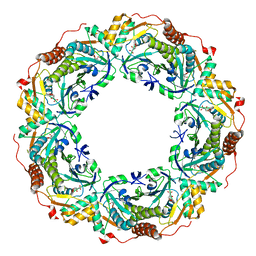 | |
6KRS
 
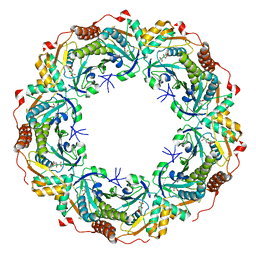 | |
8HLA
 
 | |
8HH0
 
 | |
