6IZP
 
 | | Solution structure of the complex of naphthyridine carbamate dimer and an RNA with UGGAA-UGGAA pentad | | Descriptor: | 3-[3-[(7-methyl-1,8-naphthyridin-2-yl)carbamoyloxy]propylamino]propyl ~{N}-(7-methyl-1,8-naphthyridin-2-yl)carbamate, RNA (29-MER) | | Authors: | Nagano, K, Shibata, T, Nakatani, K, Kawai, G. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Small molecule targeting r(UGGAA)n disrupts RNA foci and alleviates disease phenotype in Drosophila model
Nat Commun, 12, 2021
|
|
1RCK
 
 | |
1RCL
 
 | |
3AFB
 
 | |
1AG7
 
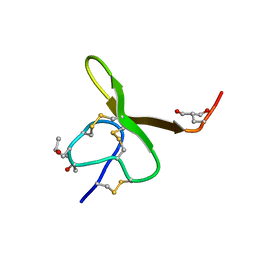 | | CONOTOXIN GS, NMR, 20 STRUCTURES | | Descriptor: | CONOTOXIN GS | | Authors: | Hill, J.M, Alewood, P.F, Craik, D.J. | | Deposit date: | 1997-04-03 | | Release date: | 1998-04-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the sodium channel antagonist conotoxin GS: a new molecular caliper for probing sodium channel geometry.
Structure, 5, 1997
|
|
3SYU
 
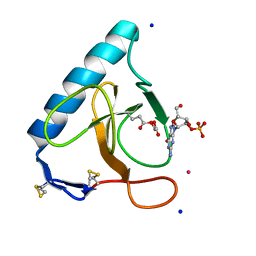 | | Re-refined coordinates for pdb entry 1det - ribonuclease T1 carboxymethylated at GLU 58 in complex with 2'GMP | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, Guanyl-specific ribonuclease T1, SODIUM ION, ... | | Authors: | Smart, O.S, Womack, T.O, Bricogne, G. | | Deposit date: | 2011-07-18 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8T8A
 
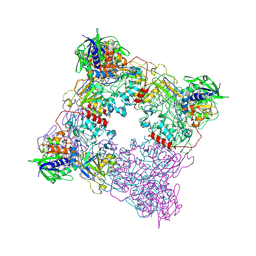 | |
8ITO
 
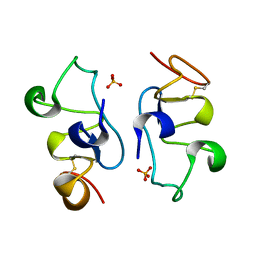 | |
8W6X
 
 | | Neutron structure of [NiFe]-hydrogenase from D. vulgaris Miyazaki F in its oxidized state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Hiromoto, T, Tamada, T. | | Deposit date: | 2023-08-30 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (1.04 Å), X-RAY DIFFRACTION | | Cite: | New insights into the oxidation process from neutron and X-ray crystal structures of an O 2 -sensitive [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
1WLE
 
 | | Crystal Structure of mammalian mitochondrial seryl-tRNA synthetase complexed with seryl-adenylate | | Descriptor: | SERYL ADENYLATE, Seryl-tRNA synthetase | | Authors: | Chimnaronk, S, Jeppesen, M.G, Suzuki, T, Nyborg, J, Watanabe, K. | | Deposit date: | 2004-06-25 | | Release date: | 2005-09-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dual-mode recognition of noncanonical tRNAs(Ser) by seryl-tRNA synthetase in mammalian mitochondria
Embo J., 24, 2005
|
|
6AKG
 
 | |
6AKF
 
 | |
6AKE
 
 | |
6IQZ
 
 | | High resolution structure of bilirubin oxidase from Myrothecium verrucaria - wild type | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bilirubin oxidase, COPPER (II) ION, ... | | Authors: | Shibata, N, Akter, M, Higuchi, Y. | | Deposit date: | 2018-11-09 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Redox Potential-Dependent Formation of an Unusual His-Trp Bond in Bilirubin Oxidase.
Chemistry, 24, 2018
|
|
6IQY
 
 | | High resolution structure of bilirubin oxidase from Myrothecium verrucaria - M467Q mutant, anaerobically prepared | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Bilirubin oxidase, ... | | Authors: | Shibata, N, Akter, M, Higuchi, Y. | | Deposit date: | 2018-11-09 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Redox Potential-Dependent Formation of an Unusual His-Trp Bond in Bilirubin Oxidase.
Chemistry, 24, 2018
|
|
6IQX
 
 | | High resolution structure of bilirubin oxidase from Myrothecium verrucaria - M467Q mutant, aerobically prepared | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Bilirubin oxidase, ... | | Authors: | Shibata, N, Akter, M, Higuchi, Y. | | Deposit date: | 2018-11-09 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Redox Potential-Dependent Formation of an Unusual His-Trp Bond in Bilirubin Oxidase.
Chemistry, 24, 2018
|
|
2YXN
 
 | | Structual basis of azido-tyrosine recognition by engineered bacterial Tyrosyl-tRNA synthetase | | Descriptor: | 3-AZIDO-L-TYROSINE, Tyrosyl-tRNA synthetase | | Authors: | Oki, K, Kobayashi, T, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-26 | | Release date: | 2008-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional replacement of the endogenous tyrosyl-tRNA synthetase-tRNATyr pair by the archaeal tyrosine pair in Escherichia coli for genetic code expansion
Nucleic Acids Res., 38, 2010
|
|
2DLC
 
 | | Crystal structure of the ternary complex of yeast tyrosyl-tRNA synthetase | | Descriptor: | MAGNESIUM ION, O-(ADENOSINE-5'-O-YL)-N-(L-TYROSYL)PHOSPHORAMIDATE, T-RNA (76-MER), ... | | Authors: | Tsunoda, M, Kusakabe, Y, Tanaka, N, Nakamura, K.T. | | Deposit date: | 2006-04-18 | | Release date: | 2007-06-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for recognition of cognate tRNA by tyrosyl-tRNA synthetase from three kingdoms.
Nucleic Acids Res., 35, 2007
|
|
3AJ4
 
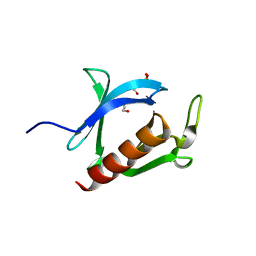 | | Crystal structure of the PH domain of Evectin-2 from human complexed with O-phospho-L-serine | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOSERINE, Pleckstrin homology domain-containing family B member 2 | | Authors: | Okazaki, S, Kato, R, Wakatsuki, S. | | Deposit date: | 2010-05-21 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Intracellular phosphatidylserine is essential for retrograde membrane traffic through endosomes
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7VEE
 
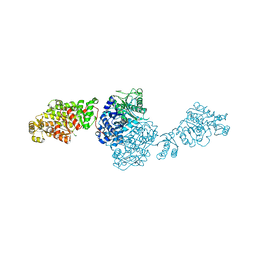 | | The ligand-free structure of GfsA KSQ-AT didomain | | Descriptor: | GLYCEROL, Polyketide synthase | | Authors: | Chisuga, T, Miyanaga, A, Nagai, A, Kudo, F, Eguchi, T. | | Deposit date: | 2021-09-08 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insight into the Reaction Mechanism of Ketosynthase-Like Decarboxylase in a Loading Module of Modular Polyketide Synthases.
Acs Chem.Biol., 17, 2022
|
|
7VEF
 
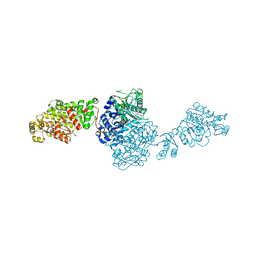 | | The structure of GfsA KSQ-AT didomain in complex with a malonate substrate analog | | Descriptor: | GLYCEROL, N-(2-acetamidoethyl)-2-nitro-ethanamide, Polyketide synthase | | Authors: | Chisuga, T, Miyanaga, A, Nagai, A, Kudo, F, Eguchi, T. | | Deposit date: | 2021-09-08 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Insight into the Reaction Mechanism of Ketosynthase-Like Decarboxylase in a Loading Module of Modular Polyketide Synthases.
Acs Chem.Biol., 17, 2022
|
|
2ZOT
 
 | |
2ZOU
 
 | | Crystal structure of human F-spondin reeler domain (fragment 2) | | Descriptor: | 1,2-ETHANEDIOL, Spondin-1 | | Authors: | Nagae, M, Nogi, T, Takagi, J. | | Deposit date: | 2008-06-07 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the F-spondin reeler domain reveals a unique beta-sandwich fold with a deformable disulfide-bonded loop
Acta Crystallogr.,Sect.D, 64, 2008
|
|
