6J4C
 
 | | Crystal structure of MarH, an epimerase for biosynthesis of Maremycins in Streptomyces, under 10 mM ZnSO4 | | Descriptor: | ACETIC ACID, Cupin superfamily protein, GLYCEROL, ... | | Authors: | Hou, Y, Liu, B, Hu, K, Zhang, R. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural basis of the mechanism of beta-methyl epimerization by enzyme MarH.
Org.Biomol.Chem., 17, 2019
|
|
6J4B
 
 | | Crystal structure of MarH, an epimerase for biosynthesis of Maremycins in Streptomyces, under 400 mM Zinc acetate | | Descriptor: | ACETIC ACID, Cupin superfamily protein, GLYCEROL, ... | | Authors: | Hou, Y, Liu, B, Hu, K, Zhang, R. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural basis of the mechanism of beta-methyl epimerization by enzyme MarH.
Org.Biomol.Chem., 17, 2019
|
|
2MVW
 
 | | Solution structure of the TRIM19 B-box1 (B1) of human promyelocytic leukemia (PML) | | Descriptor: | Protein PML, ZINC ION | | Authors: | Huang, S, Naik, M.T, Fan, P, Wang, Y, Chang, C, Huang, T. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The B-box 1 dimer of human promyelocytic leukemia protein.
J.Biomol.Nmr, 60, 2014
|
|
2K02
 
 | |
5GYH
 
 | |
5GY8
 
 | |
5GYC
 
 | |
5GYG
 
 | |
5GYA
 
 | |
5GV1
 
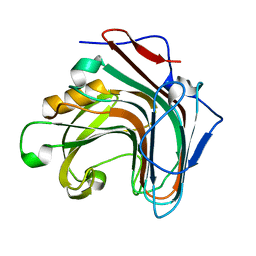 | | Crystal structure of ENZbleach xylanase wild type | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Chitnumsub, P, Jaruwat, A, Boonyapakorn, K, Noytanom, K. | | Deposit date: | 2016-09-01 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based protein engineering for thermostable and alkaliphilic enhancement of endo-beta-1,4-xylanase for applications in pulp bleaching
J. Biotechnol., 259, 2017
|
|
5GYB
 
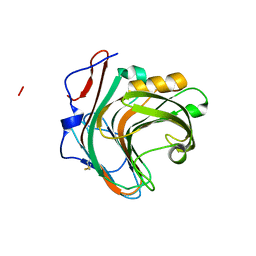 | |
5GYE
 
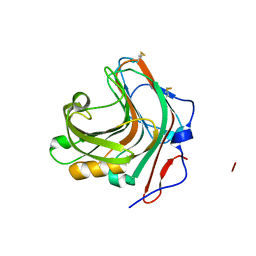 | |
5GYI
 
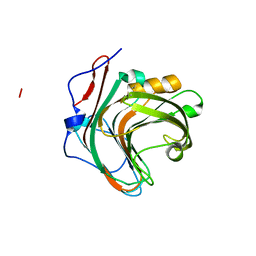 | |
5GY9
 
 | |
5GYF
 
 | |
7XMX
 
 | | Cryo-EM structure of SARS-CoV-2 spike glycoprotein in complex with three F61 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F61 heavy chain, F61 light chain, ... | | Authors: | Wang, X, Li, X. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis of a two-antibody cocktail exhibiting highly potent and broadly neutralizing activities against SARS-CoV-2 variants including diverse Omicron sublineages.
Cell Discov, 8, 2022
|
|
7XMZ
 
 | | Cryo-EM structure of SARS-CoV-2 spike glycoprotein in complex with three D2 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D2 heavy chain, D2 light chain, ... | | Authors: | Wang, X, Li, X. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis of a two-antibody cocktail exhibiting highly potent and broadly neutralizing activities against SARS-CoV-2 variants including diverse Omicron sublineages.
Cell Discov, 8, 2022
|
|
7XST
 
 | |
