1I4S
 
 | |
1JFZ
 
 | |
2K5S
 
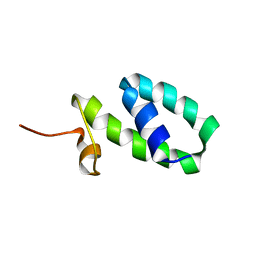 | | YmoA | | Descriptor: | Modulating protein ymoA | | Authors: | McFeeters, R.L, Byrd, R. | | Deposit date: | 2008-06-30 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The high-precision solution structure of Yersinia modulating
protein YmoA provides insight into interaction with H-NS
Biochemistry, 46, 2007
|
|
1N5B
 
 | |
1GA3
 
 | |
7KDO
 
 | | Crystal structure of Escherichia coli HPPK in complex with bisubstrate inhibitor HP-73 | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-[(2R,4R)-1-{2-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridine-6-carbonyl)amino]ethyl}-2-carboxypiperidin-4-yl]-5'-thioadenosine | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2020-10-09 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bisubstrate inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: Transition state analogs for high affinity binding.
Bioorg.Med.Chem., 29, 2021
|
|
7KDR
 
 | | Crystal structure of Escherichia coli HPPK in complex with bisubstrate inhibitor HP-75 | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-{[(2R,4R)-1-{2-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridine-6-carbonyl)amino]ethyl}-2-carboxypiperidin-4-yl]sulfonyl}-5'-deoxyadenosine, ... | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2020-10-09 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.488 Å) | | Cite: | Bisubstrate inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: Transition state analogs for high affinity binding.
Bioorg.Med.Chem., 29, 2021
|
|
