3B8M
 
 | |
1YQC
 
 | | Crystal Structure of Ureidoglycolate Hydrolase (AllA) from Escherichia coli O157:H7 | | Descriptor: | GLYOXYLIC ACID, Ureidoglycolate hydrolase | | Authors: | Raymond, S, Tocilj, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-02-01 | | Release date: | 2005-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Crystal structure of ureidoglycolate hydrolase (AllA) from Escherichia coli O157:H7
Proteins, 61, 2005
|
|
1YNF
 
 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | POTASSIUM ION, Succinylarginine dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
3ILR
 
 | | Structure of Heparinase I from Bacteroides thetaiotaomicron in complex with tetrasaccharide product | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(4-1)-4-deoxy-2-O-sulfo-beta-D-erythro-hex-4-enopyranuronic acid, 4-deoxy-2-O-sulfo-beta-D-erythro-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ... | | Authors: | Garron, M.L, Cygler, M, Shaya, D. | | Deposit date: | 2009-08-07 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural snapshots of heparin depolymerization by heparin lyase I.
J.Biol.Chem., 284, 2009
|
|
1YNH
 
 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | N~2~-(3-CARBOXYPROPANOYL)-L-ORNITHINE, POTASSIUM ION, Succinylarginine Dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M. | | Deposit date: | 2005-01-24 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
1ZPS
 
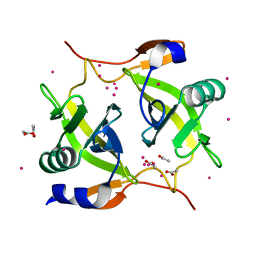 | | Crystal structure of Methanobacterium thermoautotrophicum phosphoribosyl-AMP cyclohydrolase HisI | | Descriptor: | ACETIC ACID, CADMIUM ION, Phosphoribosyl-AMP cyclohydrolase | | Authors: | Sivaraman, J, Myers, R.S, Boju, L, Sulea, T, Cygler, M, Davisson, V.J, Schrag, J.D, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Methanobacterium thermoautotrophicum Phosphoribosyl-AMP Cyclohydrolase HisI.
Biochemistry, 44, 2005
|
|
1XHN
 
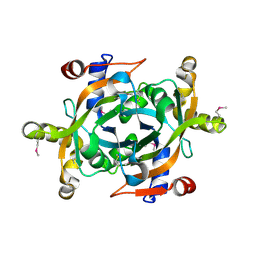 | |
3B8P
 
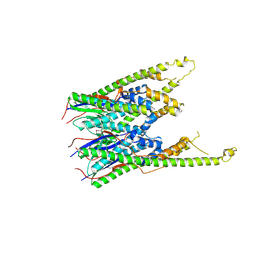 | |
2ZL1
 
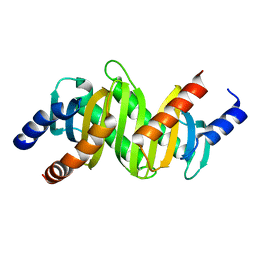 | | MP1-p14 Scaffolding complex | | Descriptor: | Mitogen-activated protein kinase kinase 1-interacting protein 1, Mitogen-activated protein-binding protein-interacting protein | | Authors: | Schrag, J.D, Cygler, M, Munger, C, Magloire, A. | | Deposit date: | 2008-04-02 | | Release date: | 2008-06-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular dynamics-solvated interaction energy studies of protein-protein interactions: the MP1-p14 scaffolding complex.
J.Mol.Biol., 379, 2008
|
|
3G2O
 
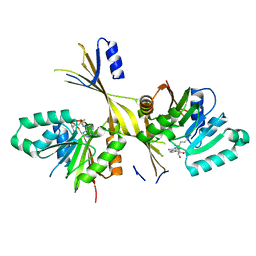 | | Crystal Structure of the Glycopeptide N-methyltransferase MtfA complexed with (S)-adenosyl-L-methionine (SAM) | | Descriptor: | PCZA361.24, S-ADENOSYLMETHIONINE | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2009-01-31 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of the glycopeptide N-methyltransferase MtfA, a tool for the biosynthesis of modified glycopeptide antibiotics.
Chem.Biol., 16, 2009
|
|
3G2M
 
 | |
3HBN
 
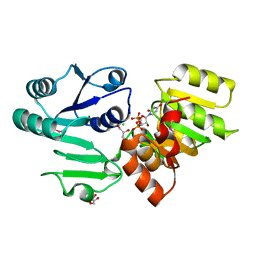 | | Crystal structure PseG-UDP complex from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, GLYCEROL, UDP-sugar hydrolase, ... | | Authors: | Rangarajan, E.S, Proteau, A, Cygler, M, Matte, A, Sulea, T, Schoenhofen, I.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional analysis of Campylobacter jejuni PseG: a udp-sugar hydrolase from the pseudaminic acid biosynthetic pathway.
J.Biol.Chem., 284, 2009
|
|
3B8N
 
 | |
3HBM
 
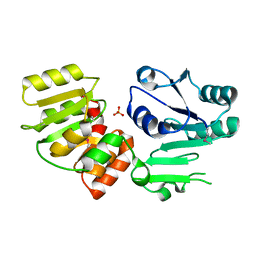 | | Crystal Structure of PseG from Campylobacter jejuni | | Descriptor: | SULFATE ION, UDP-sugar hydrolase | | Authors: | Rangarajan, E.S, Proteau, A, Cygler, M, Matte, A, Sulea, T, Schoenhofen, I.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analysis of Campylobacter jejuni PseG: a udp-sugar hydrolase from the pseudaminic acid biosynthetic pathway.
J.Biol.Chem., 284, 2009
|
|
2AHU
 
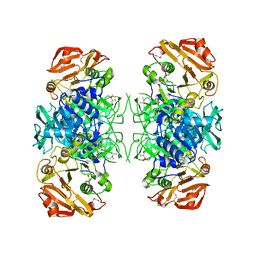 | | Crystal structure of Acyl-CoA transferase (YdiF) apoenzyme from Escherichia coli O157:H7. | | Descriptor: | putative enzyme ydiF | | Authors: | Rangarajan, E.S, Li, Y, Ajamian, E, Iannuzzi, P, Kernaghan, S.D, Fraser, M.E, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-07-28 | | Release date: | 2005-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic trapping of the glutamyl-CoA thioester intermediate of family I CoA transferases.
J.Biol.Chem., 280, 2005
|
|
3CES
 
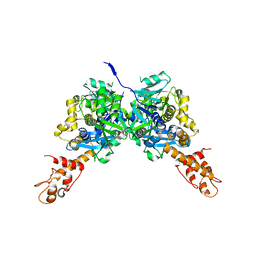 | | Crystal Structure of E.coli MnmG (GidA), a Highly-Conserved tRNA Modifying Enzyme | | Descriptor: | tRNA uridine 5-carboxymethylaminomethyl modification enzyme gidA | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2008-02-29 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Structure-function analysis of Escherichia coli MnmG (GidA), a highly conserved tRNA-modifying enzyme.
J.Bacteriol., 191, 2009
|
|
3CQK
 
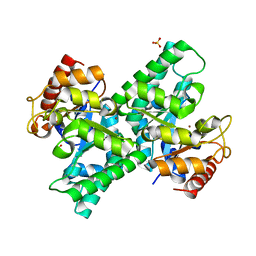 | | Crystal Structure of L-xylulose-5-phosphate 3-epimerase UlaE (form B) complex with Zn2+ and sulfate | | Descriptor: | L-ribulose-5-phosphate 3-epimerase ulaE, SULFATE ION, ZINC ION | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2008-04-03 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure of L-xylulose-5-Phosphate 3-epimerase (UlaE) from the anaerobic L-ascorbate utilization pathway of Escherichia coli: identification of a novel phosphate binding motif within a TIM barrel fold.
J.Bacteriol., 190, 2008
|
|
3CQJ
 
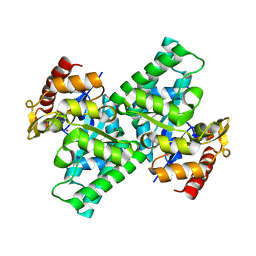 | |
3TTF
 
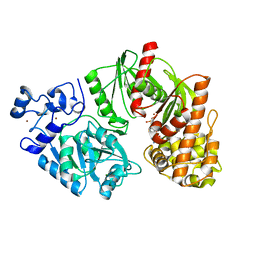 | | Crystal structure of E. coli HypF with AMP and carbamoyl phosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Transcriptional regulatory protein, ... | | Authors: | Petkun, S, Shi, R, Li, Y, Cygler, M. | | Deposit date: | 2011-09-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of Hydrogenase Maturation Protein HypF with Reaction Intermediates Shows Two Active Sites.
Structure, 19, 2011
|
|
3TSQ
 
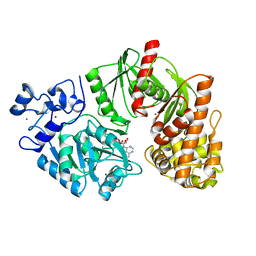 | | Crystal structure of E. coli HypF with ATP and Carbamoyl phosphate | | Descriptor: | 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, MAGNESIUM ION, Transcriptional regulatory protein, ... | | Authors: | Petkun, S, Shi, R, Li, Y, Cygler, M. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Hydrogenase Maturation Protein HypF with Reaction Intermediates Shows Two Active Sites.
Structure, 19, 2011
|
|
3TSP
 
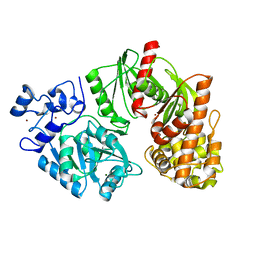 | | Crystal structure of E. coli HypF | | Descriptor: | MAGNESIUM ION, Transcriptional regulatory protein, ZINC ION | | Authors: | Petkun, S, Shi, R, Li, Y, Cygler, M. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Hydrogenase Maturation Protein HypF with Reaction Intermediates Shows Two Active Sites.
Structure, 19, 2011
|
|
6CGJ
 
 | | Structure of the HAD domain of effector protein Lem4 (lpg1101) from Legionella pneumophila | | Descriptor: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Beyrakhova, K.A, Xu, C, Cygler, M. | | Deposit date: | 2018-02-20 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Legionella pneumophilaeffector Lem4 is a membrane-associated protein tyrosine phosphatase.
J. Biol. Chem., 293, 2018
|
|
6D2C
 
 | |
6D3U
 
 | | Complex structure of Ulvan lyase from Nonlaben Ulvanivorans- NLR48 | | Descriptor: | 1,2-ETHANEDIOL, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose, CALCIUM ION, ... | | Authors: | Ulaganathan, T, Cygler, M. | | Deposit date: | 2018-04-16 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural and functional characterization of PL28 family ulvan lyase NLR48 fromNonlabens ulvanivorans.
J. Biol. Chem., 293, 2018
|
|
6DEH
 
 | | Structure of LpnE Effector Protein from Legionella pneumophila (sp. Philadelphia) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Voth, K, Chung, I.Y.W, van Straaten, K.E, Cygler, M. | | Deposit date: | 2018-05-11 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Legionella effector protein LpnE provides insights into its interaction with Oculocerebrorenal syndrome of Lowe (OCRL) protein.
FEBS J., 286, 2019
|
|
