3X0C
 
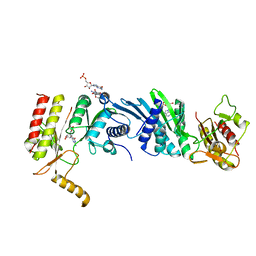 | | Crystal structure of PIP4KIIBETA I368A complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X07
 
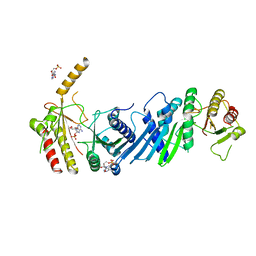 | | Crystal structure of PIP4KIIBETA N203A complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3WZZ
 
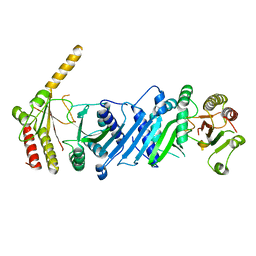 | | Crystal structure of PIP4KIIBETA | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
4B03
 
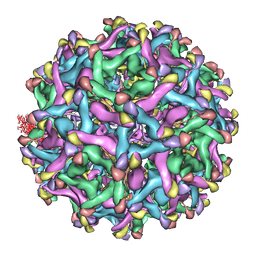 | | 6A Electron cryomicroscopy structure of immature Dengue virus serotype 1 | | Descriptor: | DENGUE VIRUS 1 E PROTEIN, DENGUE VIRUS 1 PRM PROTEIN | | Authors: | Kostyuchenko, V.A, Zhang, Q, Tan, L.C, Ng, T.S, Lok, S.M. | | Deposit date: | 2012-06-28 | | Release date: | 2013-06-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Immature and Mature Dengue Serotype 1 Virus Structures Provide Insight Into the Maturation Process.
J.Virol., 87, 2013
|
|
5Y3X
 
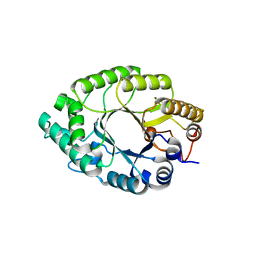 | | Crystal structure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis | | Descriptor: | Beta-xylanase | | Authors: | Liu, X, Sun, L.C, Zhang, Y.B, Liu, T.F, Xin, F.J. | | Deposit date: | 2017-07-31 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into the Thermophilic Adaption Mechanism of Endo-1,4-beta-Xylanase from Caldicellulosiruptor owensensis.
J. Agric. Food Chem., 66, 2018
|
|
5GT2
 
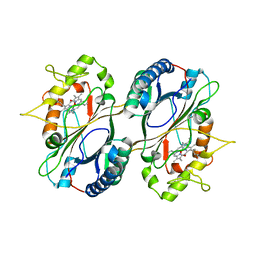 | | Crystal Structure and Biochemical Features of dye-decolorizing peroxidase YfeX from Escherichia coli O157 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Probable deferrochelatase/peroxidase YfeX | | Authors: | Ma, Y.L, Yuan, Z.G, Liu, S, Wang, J.X, Gu, L.C, Liu, X.H. | | Deposit date: | 2016-08-18 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Crystal structure and biochemical features of dye-decolorizing peroxidase YfeX from Escherichia coli O157 Asp(143) and Arg(232) play divergent roles toward different substrates
Biochem. Biophys. Res. Commun., 484, 2017
|
|
5ZO4
 
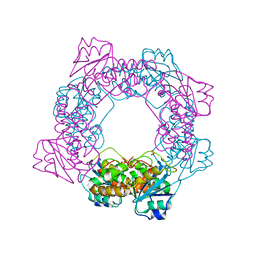 | | inactive state of the nuclease | | Descriptor: | MANGANESE (II) ION, Putative 3'-5' exonuclease family protein, SULFATE ION | | Authors: | Yuan, Z.L, Gu, L.C. | | Deposit date: | 2018-04-12 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | NrnC, an RNase D-Like Protein FromAgrobacterium, Is a Novel Octameric Nuclease That Specifically Degrades dsDNA but Leaves dsRNA Intact.
Front Microbiol, 9, 2018
|
|
5ZO3
 
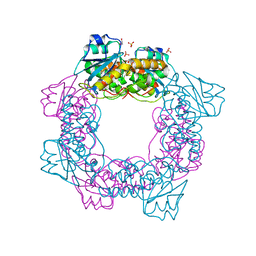 | | apo form of the nuclease | | Descriptor: | 1,2-ETHANEDIOL, Putative 3'-5' exonuclease family protein, SULFATE ION | | Authors: | Yuan, Z.L, Gu, L.C. | | Deposit date: | 2018-04-12 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.493 Å) | | Cite: | NrnC, an RNase D-Like Protein FromAgrobacterium, Is a Novel Octameric Nuclease That Specifically Degrades dsDNA but Leaves dsRNA Intact.
Front Microbiol, 9, 2018
|
|
1L9M
 
 | |
1MAJ
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | Descriptor: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | Authors: | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | Deposit date: | 1993-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
1MAK
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | Descriptor: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | Authors: | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | Deposit date: | 1993-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
1KOQ
 
 | | NEISSERIA GONORRHOEAE CARBONIC ANHYDRASE | | Descriptor: | CARBONIC ANHYDRASE, ZINC ION | | Authors: | Huang, S, Xue, Y, Chirica, L, Lindskog, S, Jonsson, B.-H. | | Deposit date: | 1998-03-22 | | Release date: | 1998-12-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of carbonic anhydrase from Neisseria gonorrhoeae and its complex with the inhibitor acetazolamide.
J.Mol.Biol., 283, 1998
|
|
1KOP
 
 | | NEISSERIA GONORRHOEAE CARBONIC ANHYDRASE | | Descriptor: | AZIDE ION, BETA-MERCAPTOETHANOL, CARBONIC ANHYDRASE, ... | | Authors: | Huang, S, Xue, Y, Chirica, L, Lindskog, S, Jonsson, B.-H. | | Deposit date: | 1998-03-22 | | Release date: | 1998-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of carbonic anhydrase from Neisseria gonorrhoeae and its complex with the inhibitor acetazolamide.
J.Mol.Biol., 283, 1998
|
|
1L9N
 
 | |
1NIF
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1NIB
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1NIE
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1AKT
 
 | | G61N OXIDIZED FLAVODOXIN MUTANT | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Mccarthy, A, Walsh, M, Higgins, T. | | Deposit date: | 1997-05-27 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of the redox potentials of FMN in Desulfovibrio vulgaris flavodoxin: thermodynamic properties and crystal structures of glycine-61 mutants.
Biochemistry, 37, 1998
|
|
1AZL
 
 | | G61V FLAVODOXIN MUTANT FROM DESULFOVIBRIO VULGARIS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Walsh, M.A, Mccarthy, A, O'Farrell, P.A, Voordouw, G, Higgins, T, Mayhew, S.G. | | Deposit date: | 1997-11-18 | | Release date: | 1998-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of the redox potentials of FMN in Desulfovibrio vulgaris flavodoxin: thermodynamic properties and crystal structures of glycine-61 mutants.
Biochemistry, 37, 1998
|
|
1B2K
 
 | | Structural effects of monovalent anions on polymorphic lysozyme crystals | | Descriptor: | IODIDE ION, PROTEIN (LYSOZYME) | | Authors: | Vaney, M.C, Broutin, I, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 1998-11-26 | | Release date: | 1998-12-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural effects of monovalent anions on polymorphic lysozyme crystals.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1A14
 
 | | COMPLEX BETWEEN NC10 ANTI-INFLUENZA VIRUS NEURAMINIDASE SINGLE CHAIN ANTIBODY WITH A 5 RESIDUE LINKER AND INFLUENZA VIRUS NEURAMINIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NC10 FV (HEAVY CHAIN), ... | | Authors: | Malby, R.L, Mccoy, A.J, Kortt, A.A, Hudson, P.J, Colman, P.M. | | Deposit date: | 1997-12-21 | | Release date: | 1998-05-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structures of single-chain Fv-neuraminidase complexes.
J.Mol.Biol., 279, 1998
|
|
1B0D
 
 | | Structural effects of monovalent anions on polymorphic lysozyme crystals | | Descriptor: | LYSOZYME, PARA-TOLUENE SULFONATE | | Authors: | Vaney, M.C, Broutin, I, Retailleau, P, Lafont, S, Hamiaux, C, Prange, T, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 1998-11-07 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural effects of monovalent anions on polymorphic lysozyme crystals.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1AKR
 
 | | G61A OXIDIZED FLAVODOXIN MUTANT | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Mccarthy, A, Walsh, M, Higgins, T. | | Deposit date: | 1997-05-27 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Modulation of the redox potentials of FMN in Desulfovibrio vulgaris flavodoxin: thermodynamic properties and crystal structures of glycine-61 mutants.
Biochemistry, 37, 1998
|
|
1AKW
 
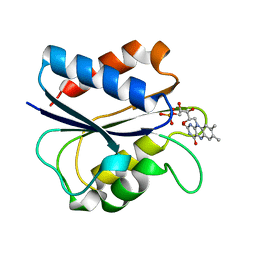 | | G61L OXIDIZED FLAVODOXIN MUTANT | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Mccarthy, A, Walsh, M, Higgins, T. | | Deposit date: | 1997-05-27 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Modulation of the redox potentials of FMN in Desulfovibrio vulgaris flavodoxin: thermodynamic properties and crystal structures of glycine-61 mutants.
Biochemistry, 37, 1998
|
|
2VNJ
 
 | |
