6WJO
 
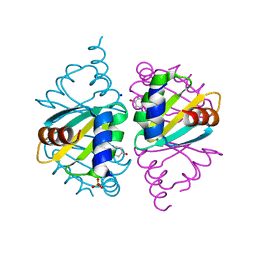 | | Crystal structure of wild-type Arginine Repressor from the pathogenic bacterium Corynebacterium pseudotuberculosis bound to tyrosine | | Descriptor: | Arginine repressor, SODIUM ION, SULFATE ION, ... | | Authors: | Nascimento, A.F.Z, Hernandez-Gonzalez, J.E, de Morais, M.A.B, Murakami, M.T, Carareto, C.M.A, Arni, R.K, Mariutti, R.B. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | A single P115Q mutation modulates specificity in the Corynebacterium pseudotuberculosis arginine repressor.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6WJP
 
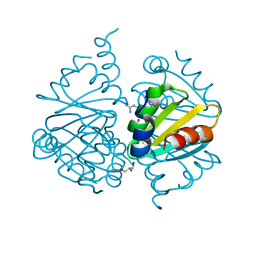 | | Crystal structure of Arginine Repressor P115Q mutant from the pathogenic bacterium Corynebacterium pseudotuberculosis bound to arginine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ARGININE, ... | | Authors: | Nascimento, A.F.Z, Hernandez-Gonzalez, J.E, de Morais, M.A.B, Murakami, M.T, Carareto, C.M.A, Arni, R.K, Mariutti, R.B. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | A single P115Q mutation modulates specificity in the Corynebacterium pseudotuberculosis arginine repressor.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6X80
 
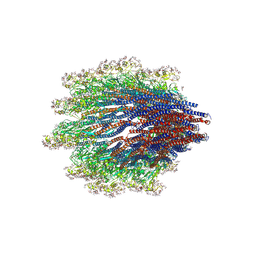 | | Structure of the Campylobacter jejuni G508A Flagellar Filament | | Descriptor: | 5,7-diamino-3,5,7,9-tetradeoxy-L-glycero-alpha-L-manno-non-2-ulopyranosonic acid, Flagellin A | | Authors: | Kreutzberger, M.A.B, Wang, F, Egelman, E.H. | | Deposit date: | 2020-06-01 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Atomic structure of the Campylobacter jejuni flagellar filament reveals how epsilon Proteobacteria escaped Toll-like receptor 5 surveillance.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4EGO
 
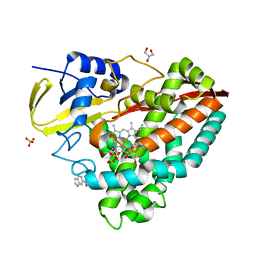 | | The X-ray crystal structure of CYP199A4 in complex with indole-6-carboxylic acid | | Descriptor: | 1H-indole-6-carboxylic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Zhou, R.M, Tan, A.B.H, Wong, L.-L. | | Deposit date: | 2012-03-31 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Investigation of the substrate range of CYP199A4: modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering
Chemistry, 18, 2012
|
|
4EGM
 
 | | The X-ray crystal structure of CYP199A4 in complex with 4-ethylbenzoic acid | | Descriptor: | 4-ethylbenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Zhou, R.M, Tan, A.B.H, Wong, L.-L. | | Deposit date: | 2012-03-31 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Investigation of the substrate range of CYP199A4: modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering
Chemistry, 18, 2012
|
|
4EGN
 
 | | The X-ray crystal structure of CYP199A4 in complex with veratric acid | | Descriptor: | 3,4-dimethoxybenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Zhou, R.M, Tan, A.B.H, Wong, L.-L. | | Deposit date: | 2012-03-31 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigation of the substrate range of CYP199A4: modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering
Chemistry, 18, 2012
|
|
4DO1
 
 | | The crystal structures of 4-methoxybenzoate bound CYP199A4 | | Descriptor: | 4-METHOXYBENZOIC ACID, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Tan, A.B.H, Zhou, R, Johnson, E.O.D, Zhang, A, Rao, Z, Wong, L.-L. | | Deposit date: | 2012-02-09 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structures of 4-methoxybenzoate bound CYP199A2 and CYP199A4: structural changes on substrate binding and the identification of an anion binding site
Dalton Trans, 41, 2012
|
|
4DNZ
 
 | | The crystal structures of CYP199A4 | | Descriptor: | CHLORIDE ION, Cytochrome P450, GLYCEROL, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Tan, A.B.H, Zhou, R, Johnson, E.O.D, Zhang, A, Rao, Z, Wong, L.-L. | | Deposit date: | 2012-02-09 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structures of 4-methoxybenzoate bound CYP199A2 and CYP199A4: structural changes on substrate binding and the identification of an anion binding site
Dalton Trans, 41, 2012
|
|
4DNJ
 
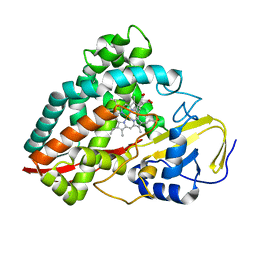 | | The crystal structures of 4-methoxybenzoate bound CYP199A2 | | Descriptor: | 4-METHOXYBENZOIC ACID, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Tan, A.B.H, Zhou, R, Johnson, E.O.D, Zhang, A, Rao, Z, Wong, L.-L. | | Deposit date: | 2012-02-08 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of 4-methoxybenzoate bound CYP199A2 and CYP199A4: structural changes on substrate binding and the identification of an anion binding site
Dalton Trans, 41, 2012
|
|
4CDU
 
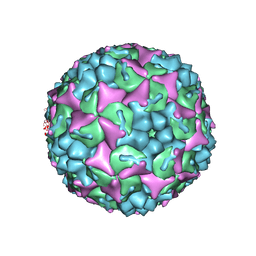 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP3 | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-06 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CDQ
 
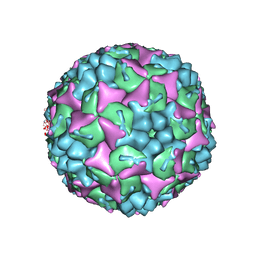 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP2 | | Descriptor: | 4-((5-(2-oxo-3-(pyridin-4-yl)imidazolidin-1-yl)pentyl)oxy)benzaldehyde O-ethyl oxime, SODIUM ION, VP1, ... | | Authors: | DeColibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-05 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4EGP
 
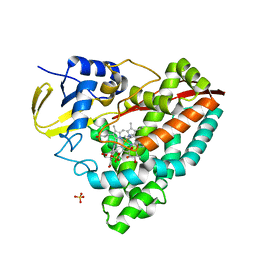 | | The X-ray crystal structure of CYP199A4 in complex with 2-naphthoic acid | | Descriptor: | CHLORIDE ION, Cytochrome P450, GLYCEROL, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Zhou, R.M, Tan, A.B.H, Wong, L.-L. | | Deposit date: | 2012-03-31 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Investigation of the substrate range of CYP199A4: modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering
Chemistry, 18, 2012
|
|
7JVI
 
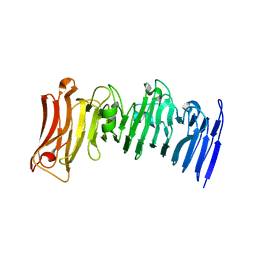 | | Crystal structure of a beta-helix domain retrieved from capybara gut metagenome | | Descriptor: | Beta-helix domain, CALCIUM ION | | Authors: | Martins, M.P, Genoroso, W.C, Domingues, M.N, Persinoti, G.F, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2020-08-21 | | Release date: | 2021-09-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Gut microbiome of the largest living rodent harbors unprecedented enzymatic systems to degrade plant polysaccharides.
Nat Commun, 13, 2022
|
|
7JVH
 
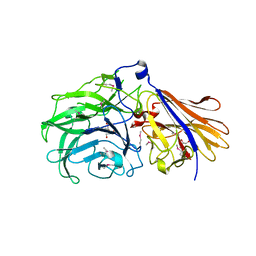 | | Crystal structure of a GH43_12 retrieved from capybara gut metagenome | | Descriptor: | GLYCEROL, Glycoside Hydrolase Family 43_12 | | Authors: | Cabral, L, Domingues, M.N, Martins, M.P, Persinoti, G.F, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2020-08-21 | | Release date: | 2021-09-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of a GH43_12 retrieved from capybara gut metagenome
To Be Published
|
|
6B7U
 
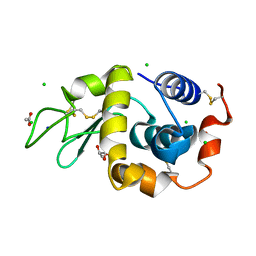 | | Structure of hen egg-white lysozyme without high-pressure pre-treatment | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Morais, M.A.B, Nascimento, A.F.Z, Tominaga, C.Y, Cristianini, M, Tribst, A.A.L, Murakami, M.T. | | Deposit date: | 2017-10-05 | | Release date: | 2018-07-25 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | How high pressure pre-treatments affect the function and structure of hen egg-white lysozyme
Innov Food Sci Emerg Technol, 47, 2018
|
|
6B7V
 
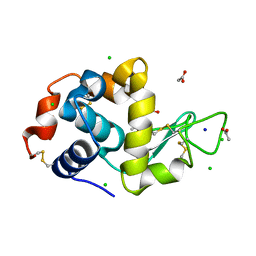 | | Structure of hen egg-white lysozyme pre-treated with high-pressure homogenization at 120 MPa | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Morais, M.A.B, Nascimento, A.F.Z, Tominaga, C.Y, Cristianini, M, Tribst, A.A.L, Murakami, M.T. | | Deposit date: | 2017-10-05 | | Release date: | 2018-07-25 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | How high pressure pre-treatments affect the function and structure of hen egg-white lysozyme
Innov Food Sci Emerg Technol, 47, 2018
|
|
6AZA
 
 | |
6BYG
 
 | | Crystal structure of the nucleophile mutant (E575A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri | | Descriptor: | Beta-mannosidase, beta-D-mannopyranose | | Authors: | Domingues, M.N, Vieira, P.S, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2017-12-20 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural basis of exo-beta-mannanase activity in the GH2 family.
J. Biol. Chem., 293, 2018
|
|
6BYC
 
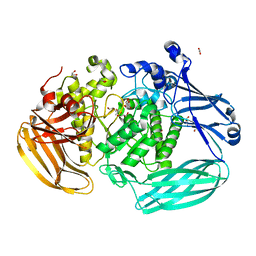 | | Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri | | Descriptor: | ACETATE ION, Beta-mannosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Domingues, M.N, Vieira, P.S, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2017-12-20 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Structural basis of exo-beta-mannanase activity in the GH2 family.
J. Biol. Chem., 293, 2018
|
|
6D25
 
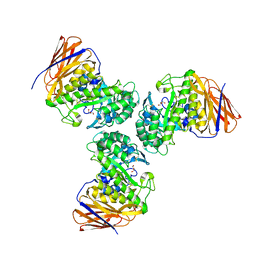 | | Crystal structure of the GH51 arabinofuranosidase from Xanthomonas axonopodis pv. citri | | Descriptor: | Alpha-L-arabinosidase, GLYCEROL | | Authors: | Santos, C.R, Morais, M.A.B, Tonoli, C.C.C, Giuseppe, P.O, Murakami, M.T. | | Deposit date: | 2018-04-13 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The mechanism by which a distinguishing arabinofuranosidase can cope with internal di-substitutions in arabinoxylans.
Biotechnol Biofuels, 11, 2018
|
|
6BYI
 
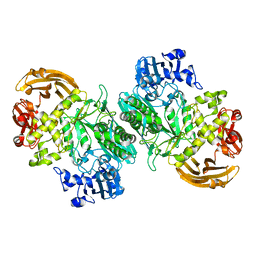 | | Crystal structure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri | | Descriptor: | Beta-mannosidase, beta-D-mannopyranose | | Authors: | Domingues, M.N, Vieira, P.S, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2017-12-20 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of exo-beta-mannanase activity in the GH2 family.
J. Biol. Chem., 293, 2018
|
|
6BYE
 
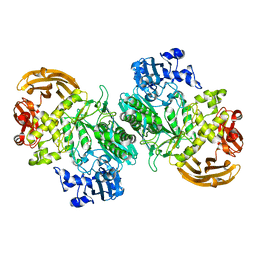 | | Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose | | Descriptor: | ACETATE ION, Beta-mannosidase, beta-D-mannopyranose | | Authors: | Domingues, M.N, Vieira, P.S, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2017-12-20 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.126 Å) | | Cite: | Structural basis of exo-beta-mannanase activity in the GH2 family.
J. Biol. Chem., 293, 2018
|
|
6B7W
 
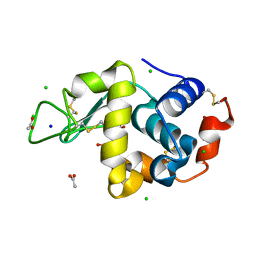 | | Structure of hen egg-white lysozyme pre-treated with high pressure (600 MPa) under isobaric condition | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Morais, M.A.B, Nascimento, A.F.Z, Tominaga, C.Y, Cristianini, M, Tribst, A.A.L, Murakami, M.T. | | Deposit date: | 2017-10-05 | | Release date: | 2018-07-25 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | How high pressure pre-treatments affect the function and structure of hen egg-white lysozyme
Innov Food Sci Emerg Technol, 47, 2018
|
|
8FOP
 
 | | Structure of Agrobacterium tumefaciens bacteriophage Milano curved tail | | Descriptor: | Tail sheath protein, Virion-associated protein | | Authors: | Sonani, R.R, Leiman, P.G, Wang, F, Kreutzberger, M.A.B, Sebastian, A, Esteves, N.C, Kelly, R.J, Scharf, B, Egelman, E.H. | | Deposit date: | 2023-01-03 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An extensive disulfide bond network prevents tail contraction in Agrobacterium tumefaciens phage Milano.
Nat Commun, 15, 2024
|
|
8GI2
 
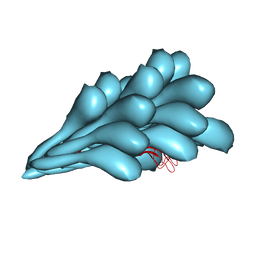 | | Cryo-EM structure of Natrinema sp. J7-2 Type IV pilus | | Descriptor: | Natrinema pilin, Orf10 | | Authors: | Sonani, R.R, Kreutzberger, M.A.B, Liu, Y, Krupovic, M, Egelman, E.H. | | Deposit date: | 2023-03-13 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The evolution of archaeal flagellar filaments.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
