7CS3
 
 | | IiPLR1 with NADP+ | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol-lariciresinol reductase | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.40021849 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CSE
 
 | | AtPrR1 with NADP+ and (-)lariciresinol | | Descriptor: | 4-[[(3S,4S,5R)-4-(hydroxymethyl)-5-(3-methoxy-4-oxidanyl-phenyl)oxolan-3-yl]methyl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44019055 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CS4
 
 | | IiPLR1 with NADP+ and (+)pinoresinol | | Descriptor: | 4-[(3S,3aR,6S,6aR)-6-(3-methoxy-4-oxidanyl-phenyl)-1,3,3a,4,6,6a-hexahydrofuro[3,4-c]furan-3-yl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol-lariciresinol reductase | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.30509257 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CS9
 
 | | AtPrR1 in apo form | | Descriptor: | Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8011415 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CSG
 
 | | AtPrR2 in apo form | | Descriptor: | Pinoresinol reductase 2 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99689388 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CSA
 
 | | AtPrR1 with NADP+ | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96212828 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
6J1M
 
 | |
8HHE
 
 | |
6J1N
 
 | | Anisodus acutangulus type III polyketide sythase AaPKS2 in complex with 4-carboxy-3-oxobutanoyl-CoA | | Descriptor: | (3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-10,14,19,21-tetraoxo-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatricosan-23-oic acid 3,5-dioxide (non-preferred name), A. acutangulus PKS2 | | Authors: | Fang, C.L, Zhang, Y. | | Deposit date: | 2018-12-28 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Tropane alkaloids biosynthesis involves an unusual type III polyketide synthase and non-enzymatic condensation.
Nat Commun, 10, 2019
|
|
6JQ1
 
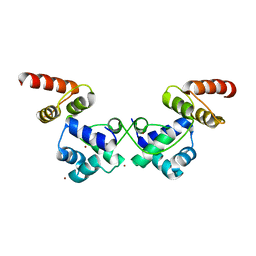 | | Crystal Structure of DdrO from Deinococcus geothermalis | | Descriptor: | LITHIUM ION, Transcriptional regulator, XRE family | | Authors: | Lu, H, Hua, Y, Zhao, Y. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and DNA damage-dependent derepression mechanism for the XRE family member DG-DdrO.
Nucleic Acids Res., 47, 2019
|
|
7VB8
 
 | |
7VMV
 
 | |
8HLG
 
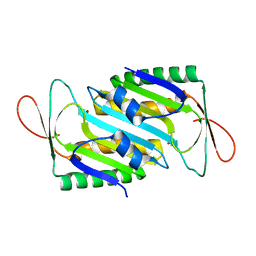 | | Crystal structure of MoaE | | Descriptor: | Molybdenum cofactor biosynthesis protein D/E, SULFATE ION | | Authors: | Cai, J, Zhao, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MoaE Is Involved in Response to Oxidative Stress in Deinococcus radiodurans.
Int J Mol Sci, 24, 2023
|
|
8IF2
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BQ.1.1 variant spike protein in complex with its receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Kimura, K, Suzuki, T, Hashiguchi, T. | | Deposit date: | 2023-02-17 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Convergent evolution of SARS-CoV-2 Omicron subvariants leading to the emergence of BQ.1.1 variant.
Nat Commun, 14, 2023
|
|
2NRV
 
 | |
7DE9
 
 | |
2NRR
 
 | |
2NRX
 
 | | Crystal structure of the C-terminal half of UvrC, in the presence of sulfate molecules | | Descriptor: | GLYCEROL, SULFATE ION, UvrABC system protein C | | Authors: | Karakas, E, Truglio, J.J, Kisker, C. | | Deposit date: | 2006-11-02 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal half of UvrC reveals an RNase H endonuclease domain with an Argonaute-like catalytic triad.
Embo J., 26, 2007
|
|
2NRT
 
 | |
6I5O
 
 | |
2NRW
 
 | |
6K2L
 
 | | Crystal structure of the Siderophore-interacting protein SipS from Aeromonas hydrophila | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Siderophore-interacting protein | | Authors: | Shang, F, Lan, J, Liu, W, Xu, Y. | | Deposit date: | 2019-05-14 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the Siderophore-interacting protein SIP from Aeromonas hydrophila.
Biochem.Biophys.Res.Commun., 519, 2019
|
|
7VQK
 
 | |
2NRZ
 
 | |
7WAS
 
 | |
