6IH2
 
 | |
5F54
 
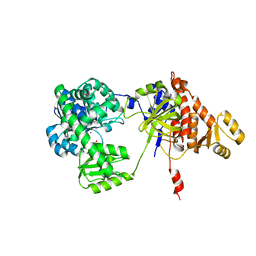 | | Structure of RecJ complexed with dTMP | | Descriptor: | MANGANESE (II) ION, Single-stranded-DNA-specific exonuclease, THYMIDINE-5'-PHOSPHATE | | Authors: | Hua, Y, Zhao, Y, Cheng, K. | | Deposit date: | 2015-12-04 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for DNA 5 -end resection by RecJ
Elife, 5, 2016
|
|
5F56
 
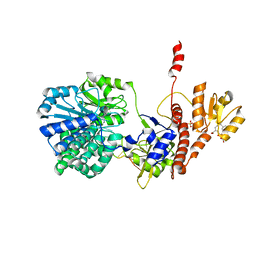 | | Structure of RecJ complexed with DNA and SSB-ct | | Descriptor: | ALA-ASP-LEU-PRO-PHE, DNA (5'-D(*CP*TP*GP*AP*TP*GP*GP*CP*A)-3'), MANGANESE (II) ION, ... | | Authors: | Zhao, Y, Hua, Y, Cheng, K. | | Deposit date: | 2015-12-04 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for DNA 5 -end resection by RecJ
Elife, 5, 2016
|
|
5B7P
 
 | |
5F55
 
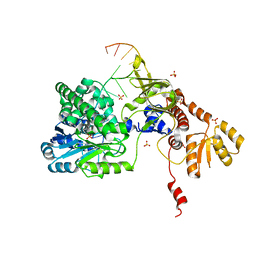 | | Structure of RecJ complexed with DNA | | Descriptor: | DNA (5'-D(*GP*AP*TP*GP*TP*AP*CP*GP*CP*TP*AP*GP*GP*C)-3'), MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Hua, Y, Zhao, Y, Cheng, K. | | Deposit date: | 2015-12-04 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for DNA 5 -end resection by RecJ
Elife, 5, 2016
|
|
5B7G
 
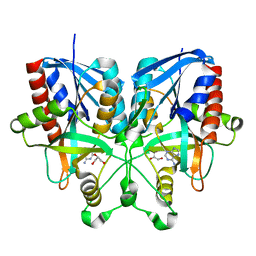 | |
6IH8
 
 | |
5B7Q
 
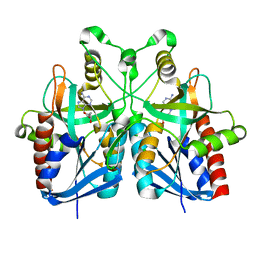 | |
5F9Q
 
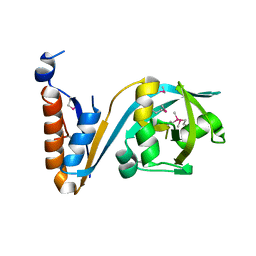 | | Crystal structure of the extracellular domain of noncanonic ABC-type transporter YknZ from Gram-positive bacteria | | Descriptor: | Macrolide export ATP-binding/permease protein YknZ | | Authors: | Xu, Y, Guo, J, Jiang, R, Jin, X, Fan, S, Quan, C.S, Ha, N.C. | | Deposit date: | 2015-12-10 | | Release date: | 2016-02-03 | | Last modified: | 2016-09-14 | | Method: | X-RAY DIFFRACTION (2.044 Å) | | Cite: | The Crystal Structure of the YknZ Extracellular Domain of ABC Transporter YknWXYZ from Bacillus amyloliquefaciens.
Plos One, 11, 2016
|
|
7R0K
 
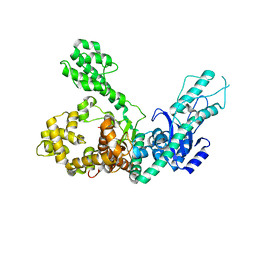 | | Crystal structure of Polymerase I from phage G20c | | Descriptor: | DNA polymerase I | | Authors: | Welin, M, Svensson, A, Hakansson, M, Al-Karadaghi, S, Linares-Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Ahlqvist, J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.972 Å) | | Cite: | Crystal structure of DNA polymerase I from Thermus phage G20c.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6J2E
 
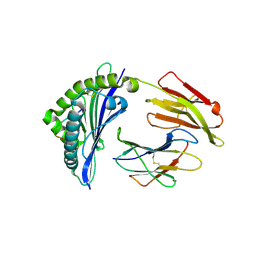 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with Ebola virus-derived peptide EBOV-NP1 | | Descriptor: | Beta-2-microglobulin, EBOV-NP1, MHC class I antigen | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
7JMN
 
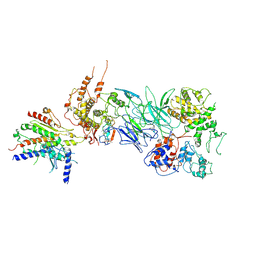 | | Tail module of Mediator complex | | Descriptor: | MED15, Mediator of RNA polymerase II transcription subunit 14, Mediator of RNA polymerase II transcription subunit 16, ... | | Authors: | Zhang, H.Q, Chen, D.C. | | Deposit date: | 2020-08-02 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Mediator structure and conformation change.
Mol.Cell, 81, 2021
|
|
6J2G
 
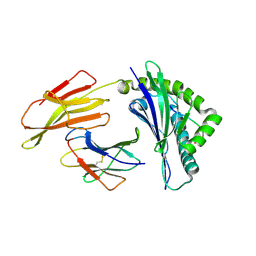 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with Ebola virus-derived peptide EBOV-NP2 | | Descriptor: | Beta-2-microglobulin, EBOV-NP2, Ptal-N*01:01 | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
6J2H
 
 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 mutant (Met52 Asp53 Leu54 deleted) in complex with Hendra virus-derived peptide HeV1 | | Descriptor: | Beta-2-microglobulin, HeV1, Ptal-N*01:01 (Met52 Asp53 Leu54 deleted) | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
6J2D
 
 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with Hendra virus-derived peptide HeV1 | | Descriptor: | HeV1, Ptal-N*01:01, beta-2 microglobulin | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
6J2F
 
 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with Hendra virus-derived peptide HeV2 | | Descriptor: | HeV2, Ptal-N*01:01, beta-2 microglobulin | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
6J2I
 
 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with H17N10 influenza-like virus-derivrd peptide H17N10-NP | | Descriptor: | Beta-2-microglobulin, H17N10-NP, Ptal-N*01:01 | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
5C0R
 
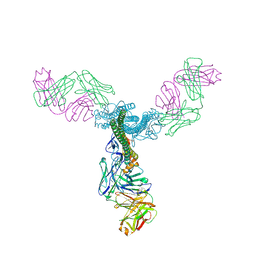 | | Crystal Structure of a Generation 3 Influenza Hemagglutinin Stabilized Stem Complexed with the Broadly Neutralizing Antibody C179 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C179 Fab heavy chain, ... | | Authors: | Boyington, J.C, kwong, P.D, Nabel, G.J, Mascola, J.R. | | Deposit date: | 2015-06-12 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.188 Å) | | Cite: | Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection.
Nat. Med., 21, 2015
|
|
7YOT
 
 | |
7YOU
 
 | |
7YOV
 
 | |
7TB8
 
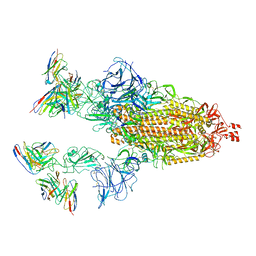 | | Cryo-EM structure of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529.
Science, 376, 2022
|
|
7TCA
 
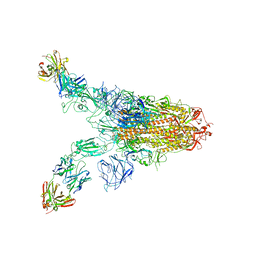 | | Cryo-EM structure of SARS-CoV-2 Omicron spike in complex with antibody A19-46.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of antibody A19-46.1, ... | | Authors: | Zhou, T, Kwong, P.D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529.
Science, 376, 2022
|
|
7TCC
 
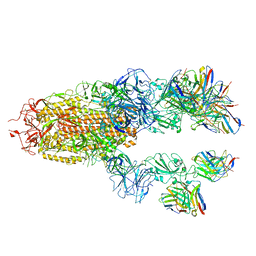 | |
7TC9
 
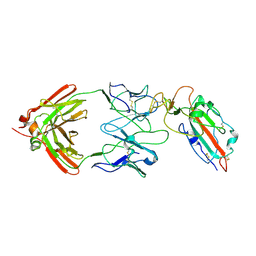 | |
