5XXN
 
 | | Crystal Structure of mutant (D286N) beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorose | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | Authors: | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahashi, Y, Sugimono, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
2CMM
 
 | | STRUCTURAL ANALYSIS OF THE MYOGLOBIN RECONSTITUTED WITH IRON PORPHINE | | Descriptor: | CYANIDE ION, MYOGLOBIN, PORPHYRIN FE(III) | | Authors: | Sato, T, Tanaka, N, Moriyama, H, Igarashi, N, Neya, S, Funasaki, N, Iizuka, T, Shiro, Y. | | Deposit date: | 1993-12-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the myoglobin reconstituted with iron porphine.
J.Biol.Chem., 268, 1993
|
|
5XXL
 
 | | Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | Authors: | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahash, Y, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
5XXO
 
 | | Crystal structure of mutant (D286N) GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorotriose | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | Authors: | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahash, Y, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
5XXM
 
 | | Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with gluconolactone | | Descriptor: | D-glucono-1,5-lactone, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | Authors: | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahash, Y, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
5YP4
 
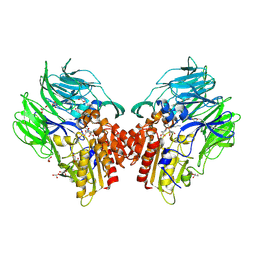 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with Lys-Pro from Pseudoxanthomonas mexicana WO24 | | Descriptor: | Dipeptidyl aminopeptidase 4, GLYCEROL, LYSINE, ... | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5YP1
 
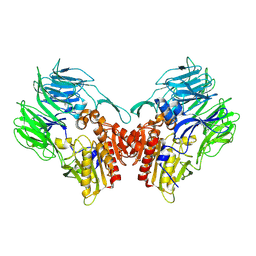 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) from Pseudoxanthomonas mexicana WO24 | | Descriptor: | Dipeptidyl aminopeptidase 4, GLYCEROL | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5YP2
 
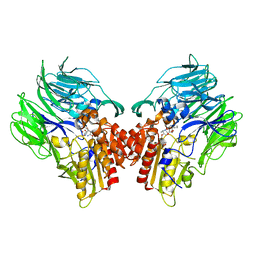 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with DPP4 inhibitor from Pseudoxanthomonas mexicana WO24 | | Descriptor: | (2S,5R)-1-[2-[[1-(hydroxymethyl)cyclopentyl]amino]ethanoyl]pyrrolidine-2,5-dicarbonitrile, Dipeptidyl aminopeptidase 4, GLYCEROL | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5YP3
 
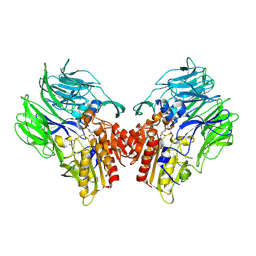 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with Ile-Pro from Pseudoxanthomonas mexicana | | Descriptor: | Dipeptidyl aminopeptidase 4, GLYCEROL, ISOLEUCINE, ... | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
1UJ0
 
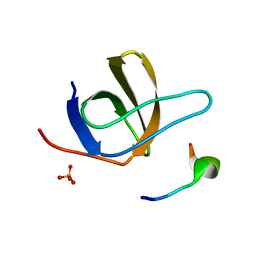 | | Crystal Structure of STAM2 SH3 domain in complex with a UBPY-derived peptide | | Descriptor: | PHOSPHATE ION, deubiquitinating enzyme UBPY, signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 | | Authors: | Kaneko, T, Kumasaka, T, Ganbe, T, Sato, T, Miyazawa, K, Kitamura, N, Tanaka, N. | | Deposit date: | 2003-07-24 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into modest binding of a non-PXXP ligand to the signal transducing adaptor molecule-2 Src homology 3 domain.
J.Biol.Chem., 278, 2003
|
|
5Z06
 
 | | Crystal structure of beta-1,2-glucanase from Parabacteroides distasonis | | Descriptor: | BDI_3064 protein, CALCIUM ION, GLYCEROL | | Authors: | Shimizu, H, Nakajima, M, Miyanaga, A, Takahashi, Y, Tanaka, N, Kobayashi, K, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization and Structural Analysis of a Novel exo-Type Enzyme Acting on beta-1,2-Glucooligosaccharides from Parabacteroides distasonis
Biochemistry, 57, 2018
|
|
1V7Z
 
 | | creatininase-product complex | | Descriptor: | MANGANESE (II) ION, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SULFATE ION, ... | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-12-26 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1XAB
 
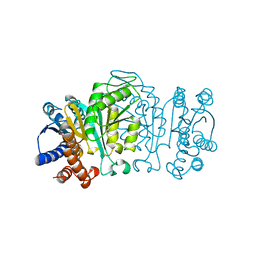 | |
1XAA
 
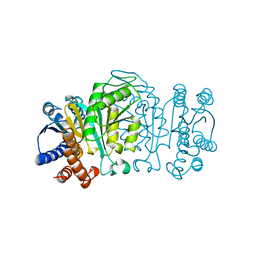 | |
1WM1
 
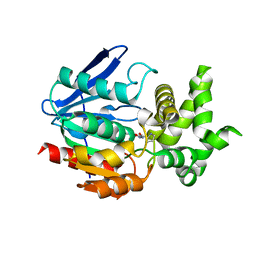 | | Crystal Structure of Prolyl Aminopeptidase, Complex with Pro-TBODA | | Descriptor: | (5-TERT-BUTYL-1,3,4-OXADIAZOL-2-YL)[(2R)-PYRROLIDIN-2-YL]METHANONE, Proline iminopeptidase | | Authors: | Nakajima, Y, Inoue, T, Ito, K, Tozaka, T, Hatakeyama, S, Tanaka, N, Nakamura, K.T, Yoshimoto, T. | | Deposit date: | 2004-07-01 | | Release date: | 2004-07-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel inhibitor for prolyl aminopeptidase from Serratia marcescens and studies on the mechanism of substrate recognition of the enzyme using the inhibitor
ARCH.BIOCHEM.BIOPHYS., 416, 2003
|
|
1WPR
 
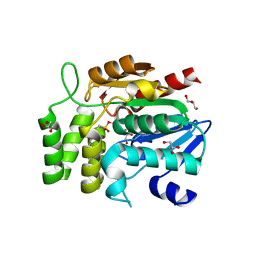 | | Crystal structure of RsbQ inhibited by PMSF | | Descriptor: | GLYCEROL, Sigma factor sigB regulation protein rsbQ, phenylmethanesulfonic acid | | Authors: | Kaneko, T, Tanaka, N, Kumasaka, T. | | Deposit date: | 2004-09-11 | | Release date: | 2005-02-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis
Protein Sci., 14, 2005
|
|
1WPW
 
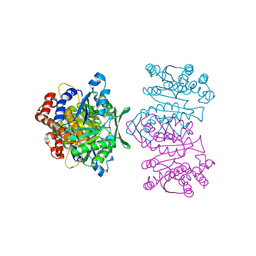 | | Crystal Structure of IPMDH from Sulfolobus tokodaii | | Descriptor: | 3-isopropylmalate dehydrogenase, MAGNESIUM ION | | Authors: | Hirose, R, Sakurai, M, Suzuki, T, Moriyama, H, Sato, T, Yamagishi, A, Oshima, T, Tanaka, N. | | Deposit date: | 2004-09-14 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of IPMDH from Sulfolobus tokodaii
To be Published
|
|
1WOJ
 
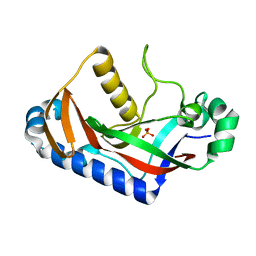 | | Crystal structure of human phosphodiesterase | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, PHOSPHATE ION | | Authors: | Sakamoto, Y, Tanaka, N, Ichimiya, T, Kurihara, T, Nakamura, K.T. | | Deposit date: | 2004-08-19 | | Release date: | 2005-03-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the catalytic fragment of human brain 2',3'-cyclic-nucleotide 3'-phosphodiesterase
J.Mol.Biol., 346, 2005
|
|
1WTM
 
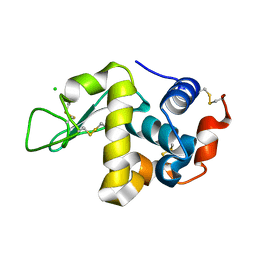 | | X-ray structure of HEW Lysozyme Orthorhombic Crystal formed in the Earth's magnetic field | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Saijo, S, Yamada, Y, Sato, T, Tanaka, N, Matsui, T, Sazaki, G, Nakajima, K, Matsuura, Y. | | Deposit date: | 2004-11-25 | | Release date: | 2004-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural consequences of hen egg-white lysozyme orthorhombic crystal growth in a high magnetic field: validation of X-ray diffraction intensity, conformational energy searching and quantitative analysis of B factors and mosaicity.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1WOM
 
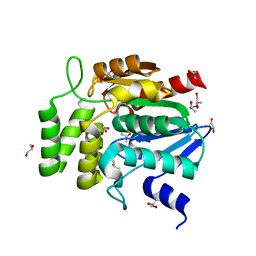 | | Crystal structure of RsbQ | | Descriptor: | MALONIC ACID, S-1,2-PROPANEDIOL, Sigma factor sigB regulation protein rsbQ | | Authors: | Kaneko, T, Kumasaka, T, Tanaka, N. | | Deposit date: | 2004-08-21 | | Release date: | 2005-02-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis
Protein Sci., 14, 2005
|
|
1UIR
 
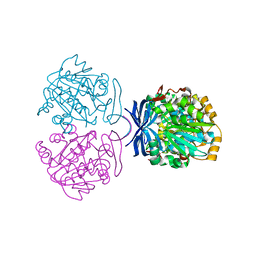 | | Crystal Structure of Polyamine Aminopropyltransfease from Thermus thermophilus | | Descriptor: | Polyamine Aminopropyltransferase | | Authors: | Ganbe, T, Ohnuma, M, Sato, T, Kumasaka, T, Oshima, T, Tanaka, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-18 | | Release date: | 2003-08-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and enzymatic properties of a triamine/agmatine aminopropyltransferase from Thermus thermophilus
J.Mol.Biol., 408, 2011
|
|
1V9I
 
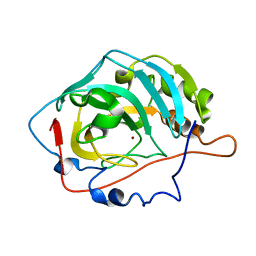 | |
1V9E
 
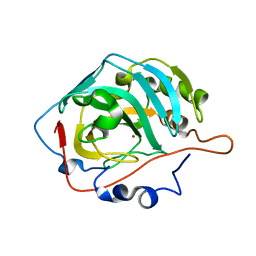 | | Crystal Structure Analysis of Bovine Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, ZINC ION | | Authors: | Saito, R, Sato, T, Ikai, A, Tanaka, N. | | Deposit date: | 2004-01-26 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of bovine carbonic anhydrase II at 1.95 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3WOR
 
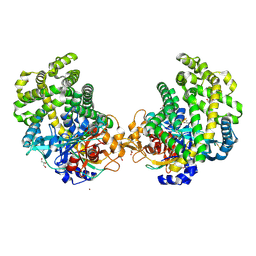 | | Crystal structure of the DAP BII octapeptide complex | | Descriptor: | Angiotensin II, GLYCEROL, ZINC ION, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOP
 
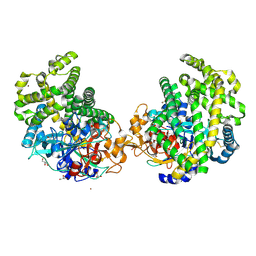 | | Crystal structure of the DAP BII hexapeptide complex II | | Descriptor: | Angiotensin IV, GLYCEROL, ZINC ION, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
