7R14
 
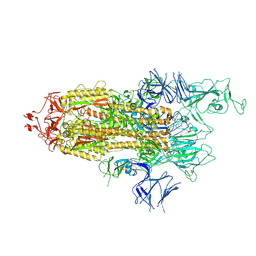 | | Alpha Variant SARS-CoV-2 Spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R1B
 
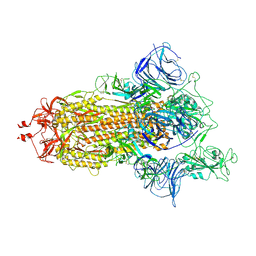 | | Mink Variant SARS-CoV-2 Spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R16
 
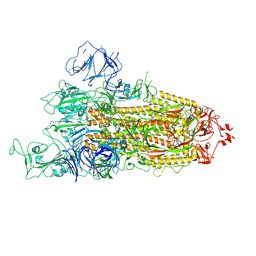 | | Beta Variant SARS-CoV-2 Spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R13
 
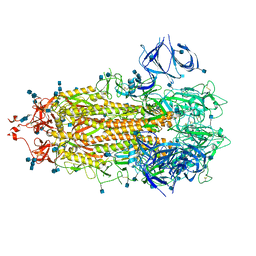 | | Alpha Variant SARS-CoV-2 Spike in Closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R19
 
 | | Mink Variant SARS-CoV-2 Spike with 2 Erect RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
3PMF
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Clark, I, Schlessman, J.L, Garcia-Moreno E, B, Heroux, A. | | Deposit date: | 2010-11-16 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7R18
 
 | | Mink Variant SARS-CoV-2 Spike in Closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R0Z
 
 | | Dissociated S1 domain of Alpha Variant SARS-CoV-2 Spike bound to ACE2 (Non-Uniform Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R12
 
 | | Dissociated S1 domain of Mink Variant SARS-CoV-2 Spike bound to ACE2 (Non-Uniform Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
3R3O
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS T62A at cryogenic temperature and with high redundancy | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2011-03-16 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6QKP
 
 | |
6QKQ
 
 | |
4E8Y
 
 | | Crystal Structure of Burkholderia cenocepacia HldA in Complex with an ATP-competitive Inhibitor | | Descriptor: | 7-O-phosphono-D-glycero-beta-D-manno-heptopyranose, CHLORIDE ION, D-beta-D-heptose 7-phosphate kinase, ... | | Authors: | Lee, T.-W, Verhey, T.B, Junop, M.S. | | Deposit date: | 2012-03-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural-functional studies of Burkholderia cenocepacia D-glycero-beta-D-manno-heptose 7-phosphate kinase (HldA) and characterization of inhibitors with antibiotic adjuvant and antivirulence properties.
J.Med.Chem., 56, 2013
|
|
4E84
 
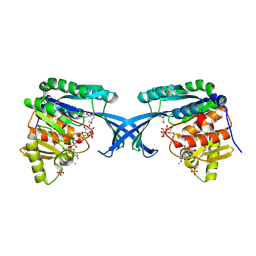 | | Crystal Structure of Burkholderia cenocepacia HldA | | Descriptor: | 1,7-di-O-phosphono-D-glycero-beta-D-manno-heptopyranose, 7-O-phosphono-D-glycero-beta-D-manno-heptopyranose, CHLORIDE ION, ... | | Authors: | Lee, T.-W, Junop, M.S. | | Deposit date: | 2012-03-19 | | Release date: | 2012-12-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural-functional studies of Burkholderia cenocepacia D-glycero-beta-D-manno-heptose 7-phosphate kinase (HldA) and characterization of inhibitors with antibiotic adjuvant and antivirulence properties.
J.Med.Chem., 56, 2013
|
|
4E8W
 
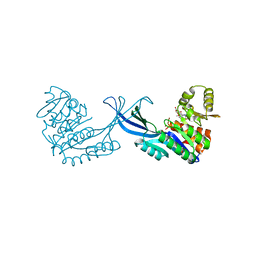 | | Crystal Structure of Burkholderia cenocepacia HldA in Complex with an ATP-competitive Inhibitor | | Descriptor: | D-beta-D-heptose 7-phosphate kinase, POTASSIUM ION, {[2-({[5-(2,6-dimethoxyphenyl)-1,2,4-triazin-3-yl]amino}methyl)-1,3-benzothiazol-5-yl]oxy}acetic acid | | Authors: | Lee, T.-W, Verhey, T.B, Junop, M.S. | | Deposit date: | 2012-03-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8654 Å) | | Cite: | Structural-functional studies of Burkholderia cenocepacia D-glycero-beta-D-manno-heptose 7-phosphate kinase (HldA) and characterization of inhibitors with antibiotic adjuvant and antivirulence properties.
J.Med.Chem., 56, 2013
|
|
4E8Z
 
 | | Crystal Structure of Burkholderia cenocepacia HldA in Complex with an ATP-competitive Inhibitor | | Descriptor: | D-beta-D-heptose 7-phosphate kinase, POTASSIUM ION, {[2-({[5-(2,6-dichlorophenyl)-1,2,4-triazin-3-yl]amino}methyl)-1,3-benzothiazol-5-yl]oxy}acetic acid | | Authors: | Lee, T.-W, Verhey, T.B, Junop, M.S. | | Deposit date: | 2012-03-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural-functional studies of Burkholderia cenocepacia D-glycero-beta-D-manno-heptose 7-phosphate kinase (HldA) and characterization of inhibitors with antibiotic adjuvant and antivirulence properties.
J.Med.Chem., 56, 2013
|
|
4AOC
 
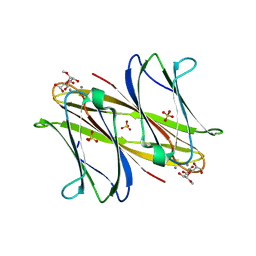 | | crystal structure of BC2L-A Lectin from Burkolderia cenocepacia in complex with methyl-heptoside | | Descriptor: | BC2L-A LECTIN, CALCIUM ION, SULFATE ION, ... | | Authors: | Marchetti, R, Malinovska, L, Lameignere, E, deCastro, C, Cioci, G, Kosma, P, Wimmerova, M, Molinaro, A, Imberty, A, Silipo, A. | | Deposit date: | 2012-03-26 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Burkholderia Cenocepacia Lectin a Binding to Heptoses from the Bacterial Lipopolysaccharide.
Glycobiology, 22, 2012
|
|
6HR5
 
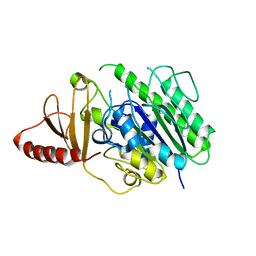 | | Structure of the S1_25 family sulfatase module of the rhamnosidase FA22250 from Formosa agariphila | | Descriptor: | Alpha-L-rhamnosidase/sulfatase (GH78), CALCIUM ION | | Authors: | Roret, T, Prechoux, A, Czjzek, M, Michel, G. | | Deposit date: | 2018-09-26 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.912 Å) | | Cite: | A marine bacterial enzymatic cascade degrades the algal polysaccharide ulvan.
Nat.Chem.Biol., 15, 2019
|
|
6HHM
 
 | | Crystal structure of the family S1_7 ulvan-specific sulfatase FA22070 from Formosa agariphila | | Descriptor: | Arylsulfatase, CALCIUM ION | | Authors: | Roret, T, Prechoux, A, Michel, G, Czjzek, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | A marine bacterial enzymatic cascade degrades the algal polysaccharide ulvan.
Nat.Chem.Biol., 15, 2019
|
|
3ZGJ
 
 | |
2KGS
 
 | | Solution structure of the amino-terminal domain of OmpATb, a pore forming protein from Mycobacterium tuberculosis | | Descriptor: | Uncharacterized protein Rv0899/MT0922 | | Authors: | Yang, Y, Auguin, D, Delbecq, S, Dumas, E, Molle, V, Saint, N. | | Deposit date: | 2009-03-18 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the Mycobacterium tuberculosis OmpATb protein: A model of an oligomeric channel in the mycobacterial cell wall
Proteins, 79, 2011
|
|
2KGW
 
 | | Solution Structure of the carboxy-terminal domain of OmpATb, a pore forming protein from Mycobacterium tuberculosis | | Descriptor: | Outer membrane protein A | | Authors: | Yang, Y, Auguin, D, Delbecq, S, Hoh, F, Dumas, E, Molle, V, Saint, N. | | Deposit date: | 2009-03-20 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the Mycobacterium tuberculosis OmpATb protein: A model of an oligomeric channel in the mycobacterial cell wall.
Proteins, 79, 2011
|
|
