2BZ1
 
 | | CRYSTAL STRUCTURE OF APO E. COLI GTP CYCLOHYDROLASE II | | Descriptor: | 2-AMINOETHANESULFONIC ACID, GLYCEROL, GTP CYCLOHYDROLASE II, ... | | Authors: | Ren, J, Kotaka, M, Lockyer, M, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2005-08-09 | | Release date: | 2005-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | GTP Cyclohydrolase II Structure and Mechanism.
J.Biol.Chem., 280, 2005
|
|
3WW6
 
 | | Crystal Structure of hen egg white lysozyme mutant N46D/D52S | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Abe, Y, Kubota, M, Ito, Y, Imoto, T, Ueda, T. | | Deposit date: | 2014-06-17 | | Release date: | 2015-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Effect on catalysis by replacement of catalytic residue from hen egg white lysozyme to Venerupis philippinarum lysozyme.
Protein Sci., 25, 2016
|
|
3ZH2
 
 | | Structure of Plasmodium falciparum lactate dehydrogenase in complex with a DNA aptamer | | Descriptor: | DNA APTAMER, L-LACTATE DEHYDROGENASE | | Authors: | Cheung, Y.W, Kwok, J, Law, A.W.L, Watt, R.M, Kotaka, M, Tanner, J.A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Discriminatory Recognition of Plasmodium Lactate Dehydrogenase by a DNA Aptamer
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4AQF
 
 | | X-ray crystallographic structure of Crimean-congo haemorrhagic fever virus nucleoprotein | | Descriptor: | NUCLEOPROTEIN, SULFATE ION | | Authors: | Wang, Y, Dutta, S, Karlberg, H, Devignot, S, Weber, F, Hao, Q, Tan, Y.J, Mirazimi, A, Kotaka, M. | | Deposit date: | 2012-04-17 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Crimean-Congo Haemorraghic Fever Virus Nucleoprotein: Superhelical Homo-Oligomers and the Role of Caspase-3 Cleavage.
J.Virol., 86, 2012
|
|
4AQG
 
 | | X-ray crystallographic structure of Crimean-congo haemorrhagic fever virus nucleoprotein | | Descriptor: | NUCLEOPROTEIN, SULFATE ION | | Authors: | Wang, Y, Dutta, S, Karlberg, H, Devignot, S, Weber, F, Hao, Q, Tan, Y.J, Mirazimi, A, Kotaka, M. | | Deposit date: | 2012-04-17 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Crimean-Congo Haemorraghic Fever Virus Nucleoprotein: Superhelical Homo-Oligomers and the Role of Caspase-3 Cleavage.
J.Virol., 86, 2012
|
|
3WW5
 
 | | Crystal Structure of hen egg white lysozyme mutant N46E/D52S | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Abe, Y, Kubota, M, Ito, Y, Imoto, T, Ueda, T. | | Deposit date: | 2014-06-17 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Effect on catalysis by replacement of catalytic residue from hen egg white lysozyme to Venerupis philippinarum lysozyme.
Protein Sci., 25, 2016
|
|
4A4D
 
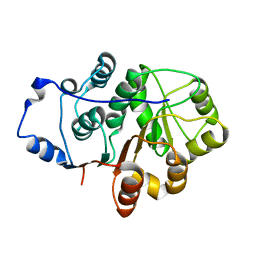 | | Crystal structure of the N-terminal domain of the Human DEAD-BOX RNA helicase DDX5 (P68) | | Descriptor: | PROBABLE ATP-DEPENDENT RNA HELICASE DDX5 | | Authors: | Dutta, S, Choi, Y.W, Kotaka, M, Fielding, B.C, Tan, Y.J. | | Deposit date: | 2011-10-11 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Variable N-Terminal Region of Ddx5 Contains Structural Elements and Auto-Inhibits its Interaction with Ns5B of Hepatitis C Virus.
Biochem.J., 446, 2012
|
|
1WQO
 
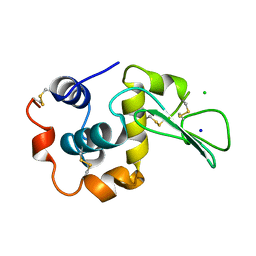 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1998-02-09 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
1WQP
 
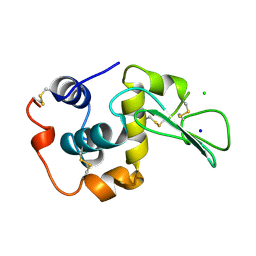 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1998-02-09 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
2AMG
 
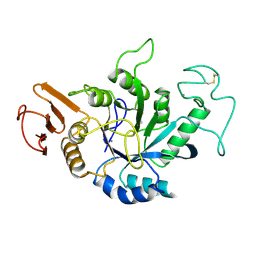 | | STRUCTURE OF HYDROLASE (GLYCOSIDASE) | | Descriptor: | 1,4-ALPHA-D-GLUCAN MALTOTETRAHYDROLASE, CALCIUM ION | | Authors: | Morishita, Y, Hasegawa, K, Matsuura, Y, Kubota, M, Sakai, S, Katsube, Y. | | Deposit date: | 1996-12-23 | | Release date: | 1997-04-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a maltotetraose-forming exo-amylase from Pseudomonas stutzeri.
J.Mol.Biol., 267, 1997
|
|
1F3P
 
 | | FERREDOXIN REDUCTASE (BPHA4)-NADH COMPLEX | | Descriptor: | FERREDOXIN REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Senda, T, Yamada, T, Sakurai, N, Kubota, M, Nishizaki, T, Masai, E, Fukuda, M, Mitsuidagger, Y. | | Deposit date: | 2000-06-06 | | Release date: | 2001-06-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of NADH-dependent ferredoxin reductase component in biphenyl dioxygenase.
J.Mol.Biol., 304, 2000
|
|
7Y8P
 
 | | Crystal structure of 4'-selenoRNA duplex | | Descriptor: | COBALT HEXAMMINE(III), RNA (5'-R(*GP*GP*AP*(IKS)P*(ILK)P*(IKS)P*GP*AP*GP*UP*CP*C)-3') | | Authors: | Kondo, J, Minakawa, N, Ohta, M, Takahashi, H, Tarashima, N. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and properties of fully-modified 4'-selenoRNA, an endonuclease-resistant RNA analog.
Bioorg.Med.Chem., 76, 2022
|
|
2BH9
 
 | | X-RAY STRUCTURE OF A DELETION VARIANT OF HUMAN GLUCOSE 6-PHOSPHATE DEHYDROGENASE COMPLEXED WITH STRUCTURAL AND COENZYME NADP | | Descriptor: | GLUCOSE-6-PHOSPHATE 1-DEHYDROGENASE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gover, S, Vandeputte-Rutten, L, Au, S.W.N, Adams, M.J. | | Deposit date: | 2005-01-08 | | Release date: | 2005-04-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of Glucose-6-Phosphate and Nadp+ Binding to Human Glucose-6-Phosphate Dehydrogenase
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1CYG
 
 | |
6JIB
 
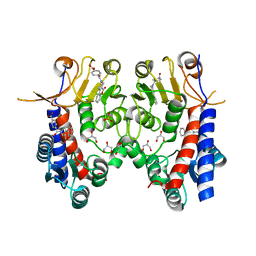 | | Human MTHFD2 in complex with DS44960156 | | Descriptor: | 4-(5-oxo-1,5-dihydro-2H-[1]benzopyrano[3,4-c]pyridine-3(4H)-carbonyl)benzoic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | Authors: | Suzuki, M, Matsui, Y, Kawai, J. | | Deposit date: | 2019-02-20 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Design and Synthesis of an Isozyme-Selective MTHFD2 Inhibitor with a Tricyclic Coumarin Scaffold.
Acs Med.Chem.Lett., 10, 2019
|
|
6JID
 
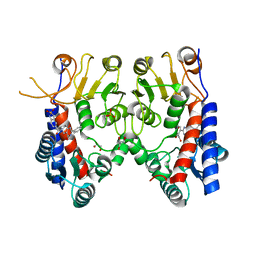 | | Human MTHFD2 in complex with Compound 1 | | Descriptor: | 5-(4-oxo-2-phenyl-1,5,7,8-tetrahydropyrido[4,3-d]pyrimidine-6(4H)-carbonyl)-1,3-dihydro-2H-2lambda~6~,1-benzothiazole-2,2-dione, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | Authors: | Suzuki, M, Matsui, Y, Matsuhashi, N, Kawai, J. | | Deposit date: | 2019-02-20 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design and Synthesis of an Isozyme-Selective MTHFD2 Inhibitor with a Tricyclic Coumarin Scaffold.
Acs Med.Chem.Lett., 10, 2019
|
|
7W51
 
 | |
7W50
 
 | |
7W4Z
 
 | |
7W52
 
 | |
7YNE
 
 | |
7DSA
 
 | |
7DS8
 
 | |
7DS4
 
 | |
7DS3
 
 | |
