5C4Y
 
 | | Crystal structure of putative TetR family transcription factor from Listeria monocytogenes | | Descriptor: | 1,2-ETHANEDIOL, Putative transcription regulator Lmo0852 | | Authors: | Chang, C, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-18 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of putative TetR family transcription factor from Listeria monocytogenes
to be published
|
|
4FX5
 
 | | von Willebrand factor type A from Catenulispora acidiphila | | Descriptor: | SODIUM ION, von Willebrand factor type A | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-07-02 | | Release date: | 2012-07-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | von Willebrand factor type A from Catenulispora acidiphila
To be Published
|
|
5BU9
 
 | | Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 | | Descriptor: | Beta-N-acetylhexosaminidase, GLYCEROL | | Authors: | Chang, C, Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-03 | | Release date: | 2015-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333
To Be Published
|
|
5BMO
 
 | | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus | | Descriptor: | ACETATE ION, POTASSIUM ION, Putative uncharacterized protein LnmX | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Cuff, M, Endres, M, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-22 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus.
to be published
|
|
4KWA
 
 | | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with choline | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, CHOLINE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with choline
To be Published
|
|
4JBE
 
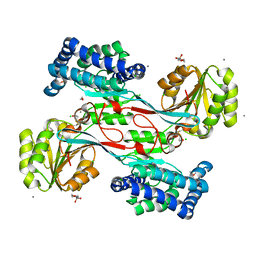 | | 1.95 Angstrom Crystal Structure of Gamma-glutamyl phosphate Reductase from Saccharomonospora viridis. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, ... | | Authors: | Minasov, G, Filippova, E.V, Halavaty, A, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of Gamma-glutamyl phosphate Reductase from Saccharomonospora viridis.
TO BE PUBLISHED
|
|
5CQF
 
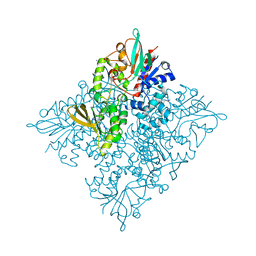 | | Crystal structure of L-lysine 6-monooxygenase from Pseudomonas syringae | | Descriptor: | IODIDE ION, L-lysine 6-monooxygenase | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Weerth, R.S, Cao, H, Yennamalli, R, Phillips Jr, G.N, Thomas, M.G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of L-lysine 6-monooxygenase from Pseudomonas syringae
To Be Published
|
|
4PIB
 
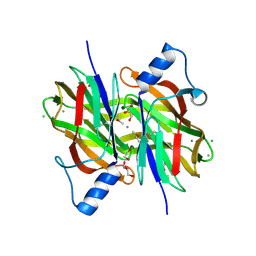 | | Crystal Structure of Uncharacterized Conserved Protein PixA from Burkholderia thailandensis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kim, Y, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-05-08 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Uncharacterized Conserved Protein PixA from Burkholderia thailandensis
To Be Published
|
|
4Q62
 
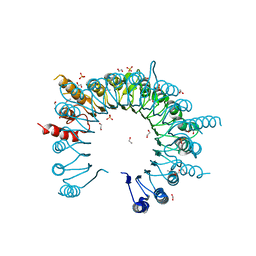 | | Crystal Structure of Leucine-rich repeat- and Coiled coil-containing Protein from Legionella pneumophila | | Descriptor: | 1,2-ETHANEDIOL, Leucine-rich repeat-and coiled coil-containing protein, SULFATE ION | | Authors: | Kim, Y, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2014-04-20 | | Release date: | 2014-05-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal Structure of Leucine-rich repeat- and Coiled coil-containing Protein from
Legionella pneumophila
To be Published
|
|
4PW4
 
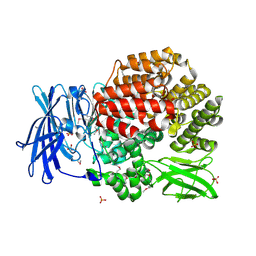 | | Crystal structure of Aminopeptidase N in complex with phosphonic acid analogue of homophenylalanine L-(R)-hPheP | | Descriptor: | Aminopeptidase N, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nocek, B, Mulligan, R, Vassiliou, S, Berlicki, L, Mucha, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-18 | | Release date: | 2014-06-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Aminopeptidase N in complex with phosphonic analogs of homophenylalanine
TO BE PUBLISHED
|
|
4Z8Z
 
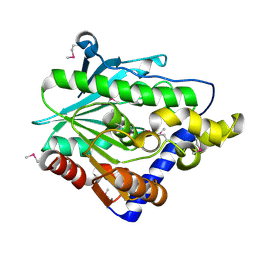 | | Crystal structure of the hypothetical protein from Ruminiclostridium thermocellum ATCC 27405 | | Descriptor: | Uncharacterized protein | | Authors: | Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Endres, M, Joachimiak, J, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-04-09 | | Release date: | 2015-05-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the hypothetical protein from Ruminiclostridium thermocellum ATCC 27405
To Be Published
|
|
4RAM
 
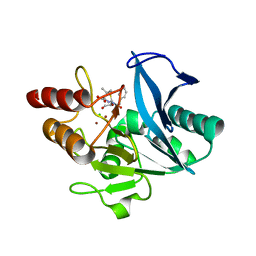 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 Mutant M67V Complexed with Hydrolyzed Penicillin G | | Descriptor: | Beta-lactamase NDM-1, CHLORIDE ION, OPEN FORM - PENICILLIN G, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2014-09-10 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.495 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 Mutant M67V Complexed with Hydrolyzed Penicillin G
To be Published
|
|
4RIT
 
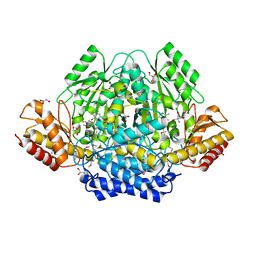 | | The yellow crystal structure of pyridoxal-dependent decarboxylase from sphaerobacter thermophilus dsm 20745 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-07 | | Release date: | 2014-10-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The yellow crystal structure of pyridoxal-dependent decarboxylase from sphaerobacter thermophilus dsm 20745
To be Published
|
|
4ZTK
 
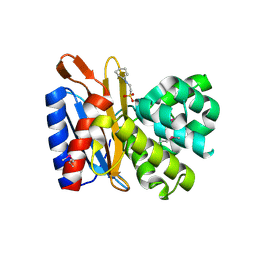 | | Transpeptidase domain of FtsI4 D,D-transpeptidase from Legionella pneumophila. | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Cell division protein FtsI/penicillin binding protein 2 | | Authors: | CUFF, M, OSIPIUK, J, WU, R, ENDRES, M, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-14 | | Release date: | 2015-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Transpeptidase domain of FtsI4 D,D-transpeptidase from Legionella pneumophila.
to be published
|
|
4JG2
 
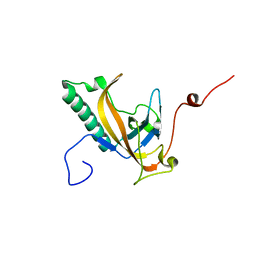 | | Structure of phage-related protein from Bacillus cereus ATCC 10987 | | Descriptor: | Phage-related protein | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-02-28 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of phage-related protein from Bacillus cereus ATCC 10987
To be Published
|
|
4ISC
 
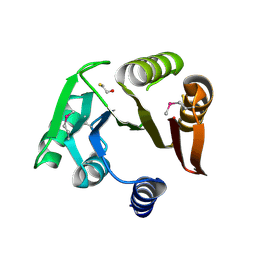 | | Crystal structure of a putative Methyltransferase from Pseudomonas syringae | | Descriptor: | BETA-MERCAPTOETHANOL, Methyltransferase | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-16 | | Release date: | 2013-02-20 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of a putative Methyltransferase from Pseudomonas syringae
To be Published
|
|
4RLG
 
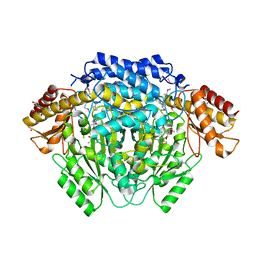 | |
4Q6J
 
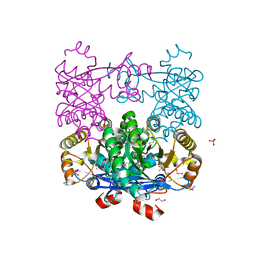 | | Crystal Structure of EAL domain Protein from Listeria monocytogenes EGD-e | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-22 | | Release date: | 2014-05-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.369 Å) | | Cite: | Crystal Structure of EAL domain Protein from Listeria monocytogenes EGD-e
To be Published
|
|
4PU2
 
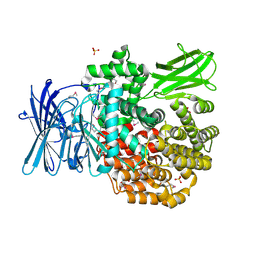 | | Crystal structure of Aminopeptidase N in complex with the phosphonic acid analogue of leucine L-(R)-LeuP | | Descriptor: | Aminopeptidase N, GLYCEROL, LEUCINE PHOSPHONIC ACID, ... | | Authors: | Nocek, B, Vassiliou, S, Berlicki, L, Mulligan, R, Mucha, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-11 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Crystal structure of Aminopeptidase N in complex with the phosphonic acid analogue of leucine
To be Published
|
|
4PDY
 
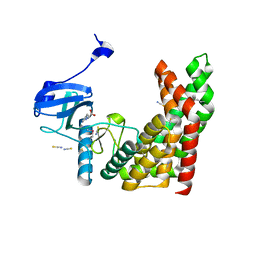 | | Crystal structure of aminoglycoside phosphotransferase from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 | | Descriptor: | Aminoglycoside phosphotransferase, HISTIDINE, THIOCYANATE ION | | Authors: | Chang, C, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-22 | | Release date: | 2014-05-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of aminoglycoside phosphotransferase from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446
To be published
|
|
4PVB
 
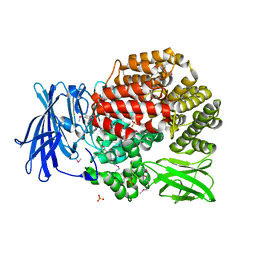 | | Crystal structure of Aminopeptidase N in complex with the phosphonic acid analogue of leucine (D-(S)-LeuP) | | Descriptor: | Aminopeptidase N, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Nocek, B, Vassiliou, S, Berlicki, L, Mulligan, R, Mucha, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-16 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Aminopeptidase N in complex with the phosphonic acid analogue of leucine (D-(S)-LeuP)
To be Published
|
|
4Q7A
 
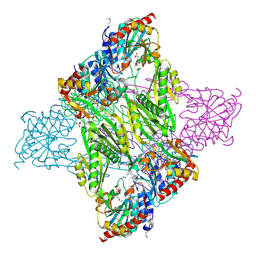 | | Crystal Structure of N-acetyl-ornithine/N-acetyl-lysine Deacetylase from Sphaerobacter thermophilus | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-ornithine/N-acetyl-lysine deacetylase, ... | | Authors: | Kim, Y, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-24 | | Release date: | 2014-07-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | Crystal Structure of N-acetyl-ornithine/N-acetyl-lysine Deacetylase from
Sphaerobacter thermophilus
To be Published
|
|
4Q63
 
 | | Crystal Structure of Legionella Uncharacterized Protein Lpg0364 | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kim, Y, Evdokimova, E, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-21 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Crystal Structure of Legionella Uncharacterized Protein Lpg0364
To be Published
|
|
4QJG
 
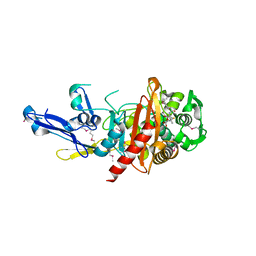 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with penicillin V | | Descriptor: | (2R,4S)-5,5-dimethyl-2-{(1R)-2-oxo-1-[(phenoxyacetyl)amino]ethyl}-1,3-thiazolidine-4-carboxylic acid, Peptidoglycan glycosyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-06-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with penicillin V
To be Published
|
|
4PYR
 
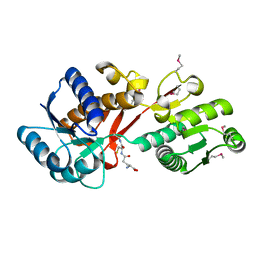 | | Structure of a putative branched-chain amino acid ABC transporter from Chromobacterium violaceum ATCC 12472 | | Descriptor: | GLUTATHIONE, Putative branched-chain amino acid ABC transporter | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-27 | | Release date: | 2014-04-23 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a putative branched-chain amino acid ABC transporter from Chromobacterium violaceum ATCC 12472
To be Published
|
|
