5LNC
 
 | |
5LPZ
 
 | |
5LPW
 
 | |
4AEC
 
 | | Crystal Structure of the Arabidopsis thaliana O-Acetyl-Serine-(Thiol)- Lyase C | | Descriptor: | ACETATE ION, CYSTEINE SYNTHASE, MITOCHONDRIAL, ... | | Authors: | Feldman-Salit, A, Wirtz, M, Lenherr, E.D, Throm, C, Hothorn, M, Scheffzek, K, Hell, R, Wade, R.C. | | Deposit date: | 2012-01-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosterically Gated Enzyme Dynamics in the Cysteine Synthase Complex Regulate Cysteine Biosynthesis in Arabidopsis Thaliana.
Structure, 20, 2012
|
|
2HQT
 
 | |
2HRA
 
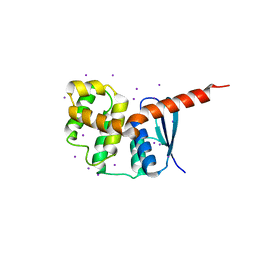 | | Crystal structures of the interacting domains from yeast glutamyl-tRNA synthetase and tRNA aminoacylation and nuclear export cofactor Arc1p reveal a novel function for an old fold | | Descriptor: | Glutamyl-tRNA synthetase, cytoplasmic, IODIDE ION | | Authors: | Simader, H, Hothorn, M, Suck, D. | | Deposit date: | 2006-07-20 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the interacting domains from yeast glutamyl-tRNA synthetase and tRNA-aminoacylation and nuclear-export cofactor Arc1p reveal a novel function for an old fold.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
3CB6
 
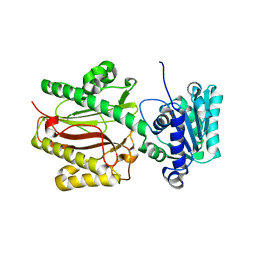 | | Crystal Structure of the S. pombe Peptidase Homology Domain of FACT complex subunit Spt16 (form B) | | Descriptor: | FACT complex subunit spt16 | | Authors: | Stuwe, T, Hothorn, M, Lejeune, E, Bortfeld-Miller, M, Scheffzek, K, Ladurner, A.G. | | Deposit date: | 2008-02-21 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The FACT Spt16 "peptidase" domain is a histone H3-H4 binding module
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2PEH
 
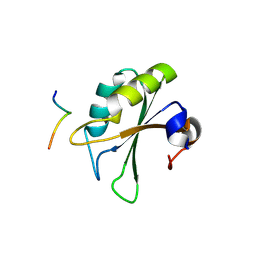 | | Crystal structure of the UHM domain of human SPF45 in complex with SF3b155-ULM5 | | Descriptor: | Splicing factor 3B subunit 1, Splicing factor 45 | | Authors: | Corsini, L, Basquin, J, Hothorn, M, Sattler, M. | | Deposit date: | 2007-04-03 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | U2AF-homology motif interactions are required for alternative splicing regulation by SPF45.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2PE8
 
 | |
5IIG
 
 | |
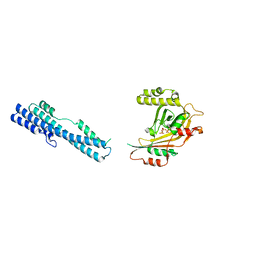
5IIT
 
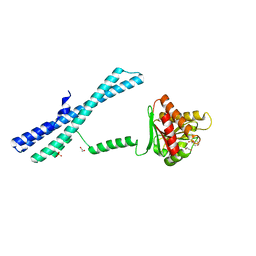 | |
5IJH
 
 | |
5IIQ
 
 | |
5IJJ
 
 | |
5IJP
 
 | |
2V9K
 
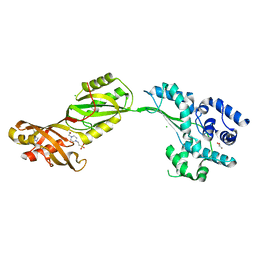 | | Crystal structure of human PUS10, a novel pseudouridine synthase. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | McCleverty, C.J, Hornsby, M, Spraggon, G, Kreusch, A. | | Deposit date: | 2007-08-23 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Pus10, a Novel Pseudouridine Synthase.
J.Mol.Biol., 373, 2007
|
|
5A66
 
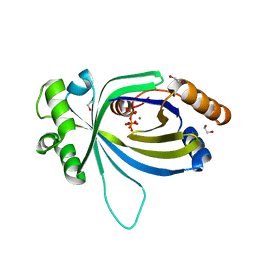 | | Crystal structure of AtTTM3 in complex with tripolyphosphate and manganese ion (form A) | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, TRIPHOSPHATE, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
6QV5
 
 | |
5A68
 
 | |
6QV7
 
 | |
6QVA
 
 | |
5A65
 
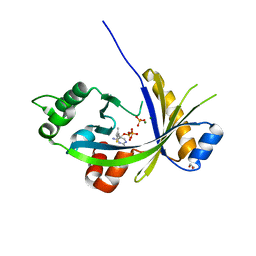 | | Crystal structure of mouse thiamine triphosphatase in complex with thiamine diphosphate, orthophosphate and magnesium ions. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
6R1H
 
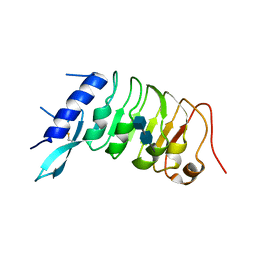 | |
5A64
 
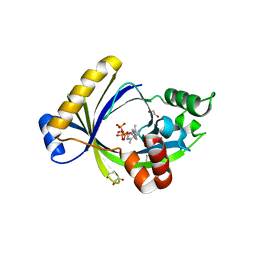 | | Crystal structure of mouse thiamine triphosphatase in complex with thiamine triphosphate. | | Descriptor: | 1,2-ETHANEDIOL, THIAMINE TRIPHOSPHATASE, TRIETHYLENE GLYCOL, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
5A60
 
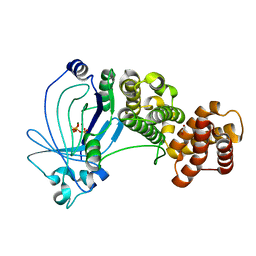 | | Crystal structure of full-length E. coli ygiF in complex with tripolyphosphate and two magnesium ions | | Descriptor: | 1,2-ETHANEDIOL, INORGANIC TRIPHOSPHATASE, MAGNESIUM ION, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
