6P58
 
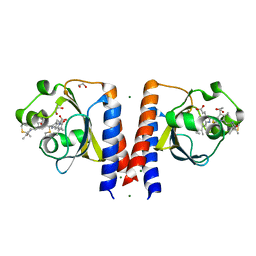 | | Dark and Steady State-Illuminated Crystal Structure of Cyanobacteriochrome Receptor PixJ at 150K | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Clinger, J.A, Miller, M.D, Buirgie, E.S, Vierstra, R.D, Phillips Jr, G.N. | | Deposit date: | 2019-05-29 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PRY
 
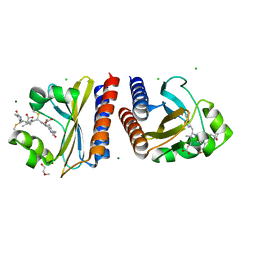 | | X-ray crystal structure of the blue-light absorbing state of PixJ from Thermosynechococcus elongatus by serial femtosecond crystallographic analysis | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Burgie, E.S, Clinger, J.A, Miller, M.D, Phillips Jr, G.N, Vierstra, R.D, Orville, A.M, Kern, J.F. | | Deposit date: | 2019-07-12 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PRU
 
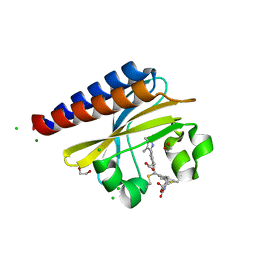 | | Photoconvertible crystals of PixJ from Thermosynechococcus elongatus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Burgie, E.S, Clinger, J.A, Miller, M.D, Phillips Jr, G.N, Vierstra, R.D. | | Deposit date: | 2019-07-11 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OIW
 
 | | Structure of Escherichia coli dGTPase bound to dGTP-1-thiol | | Descriptor: | 2'-deoxyguanosine-5'-O-(1-thiotriphosphate), Deoxyguanosinetriphosphate triphosphohydrolase, MAGNESIUM ION, ... | | Authors: | Barnes, C.O, Wu, Y, Calero, G. | | Deposit date: | 2019-04-09 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The crystal structure of dGTPase reveals the molecular basis of dGTP selectivity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OIY
 
 | | Structure of Escherichia coli bound to dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxyguanosinetriphosphate triphosphohydrolase, MANGANESE (II) ION | | Authors: | Barnes, C.O, Wu, Y, Calero, G. | | Deposit date: | 2019-04-09 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | The crystal structure of dGTPase reveals the molecular basis of dGTP selectivity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OI7
 
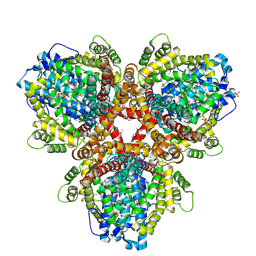 | | Se-Met structure of apo- Escherichia coli dGTPase | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, MANGANESE (II) ION, SULFATE ION | | Authors: | Calero, G, Barnes, C.O, Wu, Y. | | Deposit date: | 2019-04-08 | | Release date: | 2019-05-29 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of dGTPase reveals the molecular basis of dGTP selectivity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1N9E
 
 | | Crystal structure of Pichia pastoris Lysyl Oxidase PPLO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Guss, J.M, Duff, A.P. | | Deposit date: | 2002-11-24 | | Release date: | 2004-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of Pichia pastoris Lysyl Oxidase
Biochemistry, 42, 2003
|
|
8SZ6
 
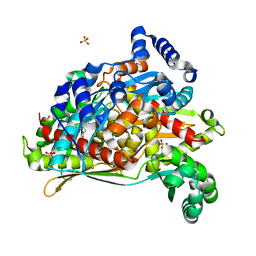 | | PmHMGR bound to mevaldehyde and CoA | | Descriptor: | (3R)-3,5,5-trihydroxy-3-methylpentanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, Mevaldyl-Coenzyme A, ... | | Authors: | Purohit, V, Stauffacher, C.V, Steussy, C.N. | | Deposit date: | 2023-05-27 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | pH-dependent reaction triggering in PmHMGR crystals for time-resolved crystallography.
Biophys.J., 123, 2024
|
|
8TSY
 
 | |
8TT0
 
 | |
8TT2
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=5.4 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8TSZ
 
 | |
8TT4
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=6.0 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8TT5
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=8.3 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8TSU
 
 | |
8TSX
 
 | |
8TT1
 
 | |
7TWR
 
 | |
7TWF
 
 | |
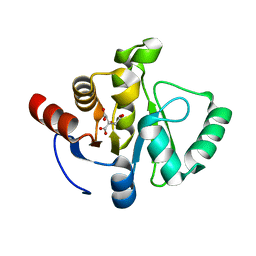
7TWP
 
 | |
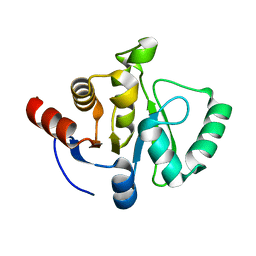
7TWH
 
 | |
7TX3
 
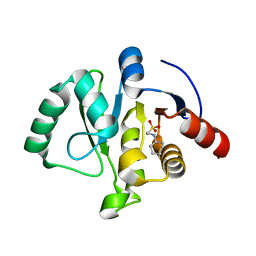 | |
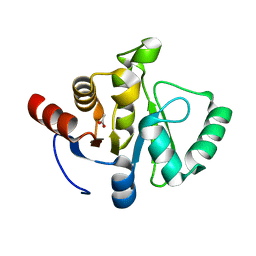
7TWN
 
 | |
7TWQ
 
 | |
7TWI
 
 | |
